 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr4g085910.1 | ||||||||
| Common Name | MTR_4g085910 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 377aa MW: 41440.8 Da PI: 10.1137 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 45 | 2.4e-14 | 302 | 357 | 5 | 60 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklk 60
+r++r++kNRe+A rsR+RK+a++ eLe +v++L++ N++L+ + e ++ ++ +
Medtr4g085910.1 302 RRQKRMIKNRESAARSRARKQAYTVELEAEVAKLKEVNEELQRKQAEFMEMQKSKE 357
79***************************************999888888877655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 3.0E-14 | 298 | 362 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.369 | 300 | 345 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.5E-12 | 302 | 357 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.88E-10 | 302 | 351 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 1.4E-13 | 302 | 351 | No hit | No description |
| CDD | cd14707 | 6.79E-27 | 302 | 356 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 305 | 320 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 377 aa Download sequence Send to blast |
MNFAWDAMSV KTPGSVNLAS QSSIYSLTFD ELQSTIGGVG KDFGSMNMDE LLKNIWNVEE 60 TQALTSLTGG GVGEGPNNPN GGTLQKQGSL TLPRTLSQRK VDEVWRDLMK DSGSSMPQRQ 120 PTLGEVTLEE FLVRAGVVKE DTPNHAQQIE RPNNNEWFSD FSRSNNNTNL LGFQQPNGNN 180 GDMSDNNNLV PKHVPLPPSS INLNHSQRPP PLFPKPTTVA FASPMHLLNN AQLGNNGRSV 240 GPGVGTLGLS ASNITAPVAS PGSKMSPDLI TKRNLDPSLL SPVPYAINRG RKCVPVEKGV 300 ERRQKRMIKN RESAARSRAR KQAYTVELEA EVAKLKEVNE ELQRKQAEFM EMQKSKEDLV 360 RTNKIKYLRR TLTGPW* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.8268 | 0.0 | leaf| root| stem | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the ABA-responsive element (ABRE). Could participate in abscisic acid-regulated gene expression. | |||||
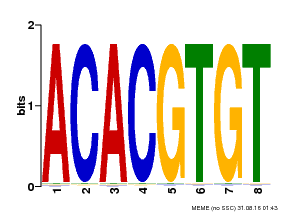
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr4g085910.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by abscisic acid (ABA) and cold. {ECO:0000269|PubMed:10636868}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT140407 | 0.0 | BT140407.1 Medicago truncatula clone JCVI-FLMt-21B4 unknown mRNA. | |||
| GenBank | BT147688 | 0.0 | BT147688.1 Medicago truncatula clone JCVI-FLMt-21J1 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003607958.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| Swissprot | Q9M7Q5 | 6e-91 | AI5L4_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 4 | ||||
| TrEMBL | G7JL68 | 0.0 | G7JL68_MEDTR; ABA response element-binding factor | ||||
| STRING | AES90155 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1962 | 34 | 81 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 4e-89 | abscisic acid responsive elements-binding factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr4g085910.1 |
| Entrez Gene | 11446220 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



