 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr4g110040.2 | ||||||||
| Common Name | MTR_4g110040 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 439aa MW: 48497.8 Da PI: 9.6088 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 55.3 | 1.2e-17 | 136 | 185 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lrel+P++ ++K +Ka+ L +++eYI+ Lq
Medtr4g110040.2 136 RSKHSATEQRRRSKINDRFQMLRELIPHS----DQKRDKASFLLEVIEYIHFLQ 185
889*************************9....9******************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.7E-24 | 131 | 207 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 5.89E-19 | 132 | 205 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.653 | 134 | 184 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.67E-15 | 136 | 189 | No hit | No description |
| Pfam | PF00010 | 3.6E-15 | 136 | 185 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.5E-13 | 140 | 190 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 439 aa Download sequence Send to blast |
MELPQTRSYG AGGAGRKSTH DFLSLYSNST AEQDPRPSAQ DSAAKLGQWT LKERTSQSFS 60 NNHHGSFNSR SSSQTTGQKN QSFMEMMKSA QDCAQDEELE NEGTFFLKRE SSNVQRGELR 120 VRVDGKSTDQ KPNTPRSKHS ATEQRRRSKI NDRFQMLREL IPHSDQKRDK ASFLLEVIEY 180 IHFLQEKVHK YEGSFQGWNN EPEKLMPWRN NDRPAESFQP RSANGGSNPS PTLLFASKVD 240 EKNITISPTT HGGIHNVESS LSTTTALKTI DHPSGITNKA FPSPISSQPN FLTSTQIGGS 300 GGAVSQLRRR LSSDAENTNY QPSVESQTMT STSEKLKEKE LTIEGGAISI SSVYSQGLLD 360 TLTQALQSSG VDLSQASISV QIELGKQASL RPNIPMPMCV SKDDEDPSKN QRKMRSGSRV 420 ASSEKSDQAL KKLKMCRS* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.14832 | 0.0 | pod| root| seed| stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
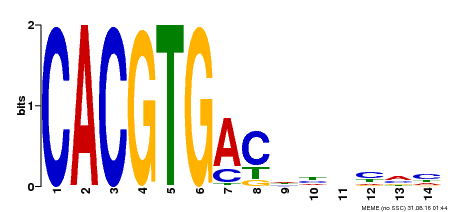
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr4g110040.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013458032.1 | 0.0 | transcription factor BIM1 isoform X2 | ||||
| TrEMBL | A0A072UQV4 | 0.0 | A0A072UQV4_MEDTR; Transcription factor BIM2-like protein | ||||
| STRING | XP_004508645.1 | 0.0 | (Cicer arietinum) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.3 | 3e-80 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr4g110040.2 |
| Entrez Gene | 25493922 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



