 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr5g006530.1 | ||||||||
| Common Name | MTR_5g006530 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 779aa MW: 85345.1 Da PI: 5.5711 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 49.9 | 6.2e-16 | 487 | 524 | 3 | 40 |
TCR 3 kkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
+k+CnCkkskClk+YCeCfaag++C e C+C++C+Nk
Medtr5g006530.1 487 CKRCNCKKSKCLKLYCECFAAGVYCIEPCSCQECFNKP 524
89**********************************96 PP
| |||||||
| 2 | TCR | 51 | 2.9e-16 | 571 | 609 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
Medtr5g006530.1 571 RHKRGCNCKKSNCLKKYCECYQGGVGCSISCRCEGCKNA 609
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 4.0E-17 | 485 | 526 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 36.811 | 486 | 611 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.5E-11 | 488 | 523 | IPR005172 | CRC domain |
| SMART | SM01114 | 5.6E-17 | 571 | 612 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 1.1E-11 | 573 | 609 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 779 aa Download sequence Send to blast |
MNMNINMDTP ERNQINTTLS KFEDSPVFNY INNLSPIKPV KSAHITQTFN SLSFSSPPSV 60 FASPHVTSLK ESRFLRRHNQ PEESKAKDSY EDVNKLYSND GAIEEPTLSR HNSRELDESA 120 EQLISVGDVS IELSNEETKF TDELPQSLQY NCGSPGYDPT LCGDEDNTLL ELSGEAVSDD 180 AYPQGGCVTD SVEAELHFQG TSQIDRKGES SDCDWDGLIP NSANMLIEAE AFKGMMQKSS 240 GSPIRLSDFV PLLQQCTRND DQKIYMVDSV ASSSQHEIEN HCSELVAATD TDHTQDNLAN 300 GSSMTGNSNE KMDNELVSVT HRGIRRRCLD FEMASVRRKI ADDNSNTNSS TQQSETMNAA 360 NENQLLPAKR NANSQRCILP GIGLHLNALA SVKDHKGKEI ENLTSGRQLS LTSSTSLLLS 420 ACQEHQHLSV VSVSISSERE LVPSDNGVQP TEDCPQPIAY MATEDFNQNS PKKKRRKSEP 480 VGEIEGCKRC NCKKSKCLKL YCECFAAGVY CIEPCSCQEC FNKPIHEDTV LQTRKQIESR 540 NPLAFAPKVI RSADSVPETG IDPNKTPASA RHKRGCNCKK SNCLKKYCEC YQGGVGCSIS 600 CRCEGCKNAF GRKDGSASIG IEGETEEETE TSDKGAEEKA IQKTEIQNIE DHPDSAGVST 660 PLRLSRSLLP MPFSSKGKPP RSFVTTLTGS GYLTSQKLAK PNSLWSQSKT FQTVPDDEMP 720 EILRGGDSSP IACIKTSSPN GKRISSPNCE MGSSPSRRGG RKLILQSIPS FPSLTHHP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 4e-17 | 488 | 608 | 12 | 120 | Protein lin-54 homolog |
| 5fd3_B | 4e-17 | 488 | 608 | 12 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 471 | 475 | KKKRR |
| 2 | 471 | 476 | KKKRRK |
| 3 | 472 | 476 | KKRRK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.7434 | 0.0 | leaf| root| stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
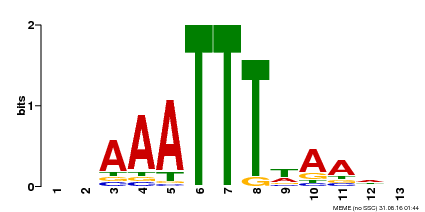
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr5g006530.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC135320 | 0.0 | AC135320.6 Medicago truncatula clone mth2-28e9, complete sequence. | |||
| GenBank | AC146852 | 0.0 | AC146852.15 Medicago truncatula clone mth2-58k21, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003610744.2 | 0.0 | protein tesmin/TSO1-like CXC 2 | ||||
| TrEMBL | G7K3P9 | 0.0 | G7K3P9_MEDTR; Putative transcription factor Tesmin family | ||||
| STRING | AES93702 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2619 | 33 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 1e-142 | TESMIN/TSO1-like CXC 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr5g006530.1 |
| Entrez Gene | 11423771 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



