 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr5g006530.2 | ||||||||
| Common Name | MTR_5g006530 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 555aa MW: 60560.2 Da PI: 7.7931 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 50.5 | 4.1e-16 | 263 | 300 | 3 | 40 |
TCR 3 kkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
+k+CnCkkskClk+YCeCfaag++C e C+C++C+Nk
Medtr5g006530.2 263 CKRCNCKKSKCLKLYCECFAAGVYCIEPCSCQECFNKP 300
89**********************************96 PP
| |||||||
| 2 | TCR | 51.6 | 1.9e-16 | 347 | 385 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN
Medtr5g006530.2 347 RHKRGCNCKKSNCLKKYCECYQGGVGCSISCRCEGCKNA 385
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 4.0E-17 | 261 | 302 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 36.811 | 262 | 387 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.0E-11 | 264 | 299 | IPR005172 | CRC domain |
| SMART | SM01114 | 5.6E-17 | 347 | 388 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 7.4E-12 | 349 | 385 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 555 aa Download sequence Send to blast |
MLIEAEAFKG MMQKSSGSPI RLSDFVPLLQ QCTRNDDQKI YMVDSVASSS QHEIENHCSE 60 LVAATDTDHT QDNLANGSSM TGNSNEKMDN ELVSVTHRGI RRRCLDFEMA SVRRKIADDN 120 SNTNSSTQQS ETMNAANENQ LLPAKRNANS QRCILPGIGL HLNALASVKD HKGKEIENLT 180 SGRQLSLTSS TSLLLSACQE HQHLSVVSVS ISSERELVPS DNGVQPTEDC PQPIAYMATE 240 DFNQNSPKKK RRKSEPVGEI EGCKRCNCKK SKCLKLYCEC FAAGVYCIEP CSCQECFNKP 300 IHEDTVLQTR KQIESRNPLA FAPKVIRSAD SVPETGIDPN KTPASARHKR GCNCKKSNCL 360 KKYCECYQGG VGCSISCRCE GCKNAFGRKD GSASIGIEGE TEEETETSDK GAEEKAIQKT 420 EIQNIEDHPD SAGVSTPLRL SRSLLPMPFS SKGKPPRSFV TTLTGSGYLT SQKLAKPNSL 480 WSQSKTFQTV PDDEMPEILR GGDSSPIACI KTSSPNGKRI SSPNCEMGSS PSRRGGRKLI 540 LQSIPSFPSL THHP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 2e-17 | 264 | 384 | 12 | 120 | Protein lin-54 homolog |
| 5fd3_B | 2e-17 | 264 | 384 | 12 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 247 | 251 | KKKRR |
| 2 | 247 | 252 | KKKRRK |
| 3 | 248 | 252 | KKRRK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.7434 | 0.0 | leaf| root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed at the first stage of pollen development in uninucleate microspores and bicellular pollen but not in tricellular and mature pollen. {ECO:0000269|PubMed:18057042}. | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous but expressed mostly in flowers with highest levels in developing ovules and microspores. {ECO:0000269|PubMed:10769244, ECO:0000269|PubMed:10769245, ECO:0000269|PubMed:18057042}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable floral-specific cell division component, required for proper organ formation in flowers. Regulates the floral meristem cell division and the inflorescence meristem organization. Plays a role in development of both male and female reproductive tissues. {ECO:0000269|PubMed:10769244, ECO:0000269|PubMed:10769245, ECO:0000269|PubMed:9043081, ECO:0000269|PubMed:9725857}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
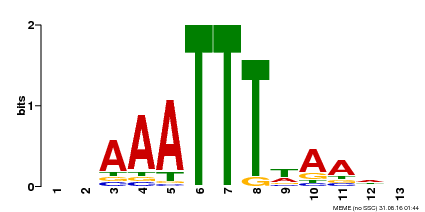
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr5g006530.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003610744.2 | 0.0 | protein tesmin/TSO1-like CXC 2 | ||||
| Swissprot | Q9LUI3 | 1e-132 | TSO1_ARATH; CRC domain-containing protein TSO1 | ||||
| TrEMBL | G7K3P9 | 0.0 | G7K3P9_MEDTR; Putative transcription factor Tesmin family | ||||
| STRING | AES93702 | 0.0 | (Medicago truncatula) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 1e-127 | TESMIN/TSO1-like CXC 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr5g006530.2 |
| Entrez Gene | 11423771 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



