 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr5g053920.1 | ||||||||
| Common Name | MTR_5g053920 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 379aa MW: 41776.1 Da PI: 9.5604 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 42.6 | 1.5e-13 | 64 | 112 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f ++eAaka++ a+++++g
Medtr5g053920.1 64 SKYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNQEDEAAKAYDVAAQRFRG 112
89****9888.8*********3.....5**********99************998 PP
| |||||||
| 2 | B3 | 99.1 | 2.7e-31 | 195 | 302 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT............---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh............ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvF 82
f+kv tpsdv+k++rlv+pk++ae+h + +++ l +ed g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F
Medtr5g053920.1 195 FEKVVTPSDVGKLNRLVIPKQHAEKHfplqkadcvqgsASAAGKGVLLNFEDIGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDTVCF 288
99*************************9888888877644555999************************************************ PP
EE-SSSEE..EEEE CS
B3 83 kldgrsefelvvkv 96
+++ e++l++ +
Medtr5g053920.1 289 QRSTGPEKQLFIDW 302
*8766666677665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 3.08E-25 | 64 | 119 | No hit | No description |
| Pfam | PF00847 | 6.4E-9 | 64 | 112 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.9E-17 | 64 | 120 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 20.02 | 65 | 120 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-20 | 65 | 120 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 7.5E-29 | 65 | 126 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 5.6E-41 | 189 | 306 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.05E-27 | 194 | 291 | No hit | No description |
| SuperFamily | SSF101936 | 4.97E-31 | 194 | 303 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 13.592 | 195 | 305 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.2E-24 | 195 | 306 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.7E-29 | 195 | 302 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 379 aa Download sequence Send to blast |
MDGGSCIDES TTTESFSITI SPTKKPSTPP PPNSLCRVGS GASAVVDSDG SGVGEAESRK 60 LPSSKYKGVV PQPNGRWGAQ IYEKHQRVWL GTFNQEDEAA KAYDVAAQRF RGKDAVTNFK 120 PLSDHNNDDM ELEFLNSHSK SEIVDMLRKH TYNDELEQSK RSHGFVSRRS HGCSDSVNFA 180 SSAYNTDKKA REALFEKVVT PSDVGKLNRL VIPKQHAEKH FPLQKADCVQ GSASAAGKGV 240 LLNFEDIGGK VWRFRYSYWN SSQSYVLTKG WSRFVKEKNL KAGDTVCFQR STGPEKQLFI 300 DWKARKNVNE VGLFVPVGPV VEPVQMVRLF GVNILKQLPG SDVNSNGNVI AGFCNGKRKE 360 LNMFTFDSCK KPKIIGAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 5e-55 | 192 | 305 | 11 | 117 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 356 | 372 | KRKELNMFTFDSCKKPK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.12539 | 0.0 | root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in leaves. {ECO:0000269|PubMed:18718758}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences. Functionally redundant with TEM2. {ECO:0000269|PubMed:18718758}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00024 | PBM | Transfer from AT1G25560 | Download |
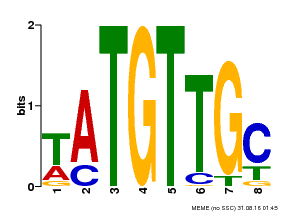
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr5g053920.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Expressed with a circadian rhythm showing a peak at dawn. {ECO:0000269|PubMed:18718758}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CU914134 | 0.0 | CU914134.2 Medicago truncatula chromosome 5 clone mte1-13k8, COMPLETE SEQUENCE. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003614455.2 | 0.0 | AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| Swissprot | Q9C6M5 | 1e-138 | RAVL1_ARATH; AP2/ERF and B3 domain-containing transcription repressor TEM1 | ||||
| TrEMBL | A0A396HTJ6 | 0.0 | A0A396HTJ6_MEDTR; Putative transcription factor RAV family | ||||
| TrEMBL | G7JY94 | 0.0 | G7JY94_MEDTR; AP2/ERF and B3 domain transcription factor | ||||
| STRING | AES97413 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2920 | 32 | 74 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25560.1 | 1e-136 | RAV family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr5g053920.1 |
| Entrez Gene | 11436458 |



