 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr5g076270.1 | ||||||||
| Common Name | MTR_5g076270 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 842aa MW: 93282.7 Da PI: 6.3571 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.8 | 2.1e-23 | 128 | 229 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f+k+lt sd++++g +++p++ ae+ +++++ ++l+ +d++ +W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F ++
Medtr5g076270.1 128 FCKTLTASDTSTHGGFSVPRRAAEKVfppldfsQQPPA--QELIARDLHDVEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI--WN 217
99*****************************8544444..48************************************************..88 PP
SEE..EEEEE-S CS
B3 88 sefelvvkvfrk 99
++ +l+++++r+
Medtr5g076270.1 218 EKNQLLLGIRRA 229
99999****997 PP
| |||||||
| 2 | Auxin_resp | 118.3 | 6.3e-39 | 254 | 337 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+t+s F+v++nPras+seFv++++k+ ka++ ++vsvGmRf+m fete+ss rr++Gt++++sd+dpvrWpnS+Wrs+k
Medtr5g076270.1 254 AAHAAATNSCFTVFFNPRASPSEFVIPLSKYIKAVYhTRVSVGMRFRMLFETEESSVRRYMGTITSISDMDPVRWPNSHWRSVK 337
79*********************************9789******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 2.75E-49 | 112 | 257 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 4.1E-42 | 121 | 243 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.58E-21 | 127 | 228 | No hit | No description |
| PROSITE profile | PS50863 | 12.379 | 128 | 230 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 3.1E-24 | 128 | 230 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.2E-21 | 128 | 229 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 6.1E-34 | 254 | 337 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 3.5E-8 | 707 | 799 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 4.56E-7 | 711 | 789 | No hit | No description |
| PROSITE profile | PS51745 | 24.483 | 715 | 799 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 842 aa Download sequence Send to blast |
MKLSTSGMSQ QGHEGGEKKC LNSELWHACA GPLVSLPTAG TRVVYFPQGH SEQVSATTNR 60 EIDGQIPNYP SLPPQLICQL HNVTMHADVE TDEVYAQMTL QPLTPQEQKD TFLPMELGIP 120 SKQPTNYFCK TLTASDTSTH GGFSVPRRAA EKVFPPLDFS QQPPAQELIA RDLHDVEWKF 180 RHIFRGQPKR HLLTTGWSVF VSAKRLVAGD SVLFIWNEKN QLLLGIRRAS RPQTVMPSSV 240 LSSDSMHIGL LAAAAHAAAT NSCFTVFFNP RASPSEFVIP LSKYIKAVYH TRVSVGMRFR 300 MLFETEESSV RRYMGTITSI SDMDPVRWPN SHWRSVKVGW DESTAGERQP RVSLWEIEPL 360 TTFPMYPSLF PLRLKRPWHP GTSSFLDGRD EATNGLMWMR GGPGDHGLNA MNFQGAGLLP 420 WMQPRLDPTL LGNDHNQQYQ AMLAAAGLQN QGNVDLLRQQ MMNFQQPFNY QQSGNLSPMQ 480 LQQQQAIQQS VSTNNIMQPQ GQGLAENLSQ HILQKSHNNR ENQTQQHSYQ DSVLIQGDPL 540 HQKQHSSLPS PSYTKPDFID SGMKFTASVS PGQNMLGSLS SEGSGNLLNL SRSGHSMLTE 600 QSPQQSWASK YSPSQVDAIG NSMSHVQYSG RDTSIVPPHC SSDAQNSVLF GVNIDSSGLL 660 LPTTVPRYTT ASAHADASTM PLGESSFQGS PYPCMQDSSE LLQSAGQVDA QNQTPIFVKV 720 YKSGSVGRSL DISRFNSYHE LREELAQMFG IEGKFEDPLR SGWQLVFVDR ENDVLLLGDD 780 PWESFVNNVW YIKILSPEDI QKMGEEAIES LGPSSGQRMN NTGAESHDIV SGLPSLGSLE 840 Y* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-159 | 15 | 358 | 45 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.7756 | 0.0 | leaf| pod| root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in sepals at stages 11, 12 and 13 of flower development. Highly expressed in petals at stages 9-10, decreases at stage 11 and disappears after flower stage 12. In anthers, expressed at stage 11 in the tapetum, disappears early in stage 12 when the tapetum degrades and reappears throughout the anther late in stage 12 to persist at least until stage 13. In stamen filaments, expressed at stages 12 to 13, especially near the apical end of the filament. Expressed throughout the gynoecium at early stages up to stage 12, especially strongly in ovules. Expression in gynoecium decreases late in stage 12, but persists through stage 13, especially near the apical end including the style. Expressed in the mesocarp of the fruit and the carpel septum during fruit growth. {ECO:0000269|PubMed:16107481, ECO:0000269|PubMed:16829592}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant. {ECO:0000269|PubMed:10476078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Regulates both stamen and gynoecium maturation. Promotes jasmonic acid production. Partially redundant with ARF6. Involved in fruit initiation. Acts as an inhibitor to stop further carpel development in the absence of fertilization and the generation of signals required to initiate fruit and seed development. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16107481, ECO:0000269|PubMed:16829592}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
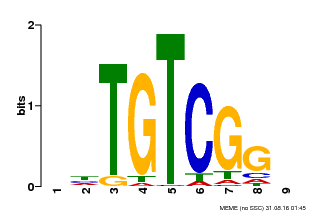
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr5g076270.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR167. {ECO:0000269|PubMed:17021043}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CU326393 | 0.0 | CU326393.2 Medicago truncatula chromosome 5 clone mth4-5l6, COMPLETE SEQUENCE. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003616115.1 | 0.0 | auxin response factor 8 | ||||
| Swissprot | Q9FGV1 | 0.0 | ARFH_ARATH; Auxin response factor 8 | ||||
| TrEMBL | G7KFN6 | 0.0 | G7KFN6_MEDTR; Auxin response factor | ||||
| STRING | AES99073 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4958 | 28 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G37020.1 | 0.0 | auxin response factor 8 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr5g076270.1 |
| Entrez Gene | 11432564 |



