 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr7g053410.2 | ||||||||
| Common Name | MTR_7g053410 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 536aa MW: 58841.2 Da PI: 6.8492 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 28.1 | 3.5e-09 | 327 | 374 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L++l+P + +k Ka +L + ++Y++sLq
Medtr7g053410.2 327 SHSLAERVRREKISERMKLLQDLVPGC----NKVTGKALMLDEIINYVQSLQ 374
8*************************9....677*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.72E-10 | 321 | 378 | No hit | No description |
| SuperFamily | SSF47459 | 7.07E-17 | 321 | 392 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 9.2E-17 | 323 | 391 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.663 | 323 | 373 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.9E-6 | 327 | 374 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.6E-10 | 329 | 379 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 536 aa Download sequence Send to blast |
MENQFFSNNH FDQSQPSSMP TWQSISSEQQ NQDCFYTPID QCVNQFDSAL SSMVSSPAAS 60 NSNMSNDNFV IRELIGKLGN IGNCSDEISQ TLVAANSYIN SNNNTSCYNT PLNSPPKLNI 120 STMAMNSTVA EFSTDPGFAE RAAKFSCFGS RSFNGRTTQL SQSQRSTPLL ENGKLSRVSS 180 SPSLKELGSQ SQIGIQENMN CSTQLQDQIE LNNSQEESTI SENGVKPSPY VNTRKRKVSS 240 KGKTKETSTS SNPPMACEAE DSNAKRMKTN EGEKIENGKV KAEDESKGGT NSNNGGDEKQ 300 NKSNSKPPEA PKDYIHVRAR RGQATDSHSL AERVRREKIS ERMKLLQDLV PGCNKVTGKA 360 LMLDEIINYV QSLQRQVEFL SMKLATVNTR VDFSIESLIS KDMFQSNNSL AHPIFPLDSS 420 APSYYGQQQQ QNPAIHNNIS NGTVTHNSVD PLDGGLCQNL GMHLSSLSGF NEAGSQYPLT 480 FSEDDLNTIV QMGFGQTDNR KTPIQFQNLN GNKPNIKKVY VNVISFSIGL IKKLC* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.16210 | 0.0 | root| stem | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00660 | PBM | Transfer from Pp3c6_9520 | Download |
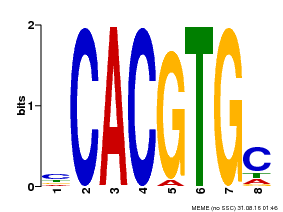
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr7g053410.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013448523.1 | 0.0 | transcription factor bHLH62 | ||||
| TrEMBL | A0A072TZD2 | 0.0 | A0A072TZD2_MEDTR; BHLH transcription factor | ||||
| STRING | XP_004492265.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2530 | 34 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G07340.1 | 1e-106 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr7g053410.2 |
| Entrez Gene | 25498190 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



