 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Medtr8g100050.1 | ||||||||
| Common Name | MTR_8g100050 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Trifolieae; Medicago
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 834aa MW: 93075.2 Da PI: 6.4859 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 75.8 | 4.9e-24 | 162 | 263 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++ +++a+e+ + k++ +++l+ +d++g++W++++i+r++++r++l++GW+ Fv++++L +gD ++F + +
Medtr8g100050.1 162 FCKTLTASDTSTHGGFSVLRRHADEClppldmS-KQPPTQELVAKDLHGNEWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL--RGE 252
99*****************************63.4445569************************************************..449 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
++el+v+v+r+
Medtr8g100050.1 253 NGELRVGVRRA 263
999*****996 PP
| |||||||
| 2 | Auxin_resp | 117.6 | 1e-38 | 288 | 370 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+ t+++F+v+Y+Pr+s++eF+v++++++++lk+++++GmRfkm+fe+e+++e+r++Gt+vg++d+d+ rWp SkWr+Lk
Medtr8g100050.1 288 AWHAVLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGTIVGIEDSDSKRWPTSKWRCLK 370
79*******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.31E-40 | 158 | 291 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 9.8E-42 | 158 | 277 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 7.43E-22 | 160 | 262 | No hit | No description |
| Pfam | PF02362 | 5.5E-22 | 162 | 263 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.8E-23 | 162 | 264 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.026 | 162 | 264 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 2.5E-36 | 288 | 370 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 6.3E-10 | 623 | 798 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 22.791 | 710 | 792 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 7.36E-10 | 713 | 787 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 834 aa Download sequence Send to blast |
MFGMRDLEEE RNMASSEVSM KGNSVNGKGE NNVDGVGDAQ NGSSSSSTGR EAEAALYREL 60 WHACAGPLVT VPREGELVFY FPQGHIEQVE ASTNQASEQH MPVYDLRPKI LCRVINVMLK 120 AEPDTDEVFA QVTLVPEPNQ DENAVEKEAP PAPPPRFHVH SFCKTLTASD TSTHGGFSVL 180 RRHADECLPP LDMSKQPPTQ ELVAKDLHGN EWRFRHIFRG QPRRHLLQSG WSVFVSSKRL 240 VAGDAFIFLR GENGELRVGV RRAMRQQGNV PSSVISSHSM HLGVLATAWH AVLTGTMFTV 300 YYKPRTSPAE FIVPYDQYME SLKNNYTIGM RFKMRFEGEE APEQRFTGTI VGIEDSDSKR 360 WPTSKWRCLK VRWDETSNIP RPERVSPWKI EPALAPPALN PLPMPRPKRP RANVVPSSPD 420 SSVLTREASS KVSMDPLPTS GFQRVLQGQE SSTLRGNLAE SNDSYTAEKS VAWTPATDEE 480 KMDAVSTSRR YGSENWMPMS RQEPTYSDLL SGFGSTREGK HNMLTQWPVM PPGLSLNFLH 540 SNMKGSAQGS DNATYQAQGN MRYSAFGDYS VLHGHKVENP HGNFLMPPPP PTQYESPHSR 600 ELSQKQMSAK ISEAAKPKDS DCKLFGFSLL SSPTMLEPSL SQRNATSETS SHMQISSQHH 660 TFENDQKSEH SKSSKPADKL VIVDEHEKQL QTSQPHVKDV QLKPQSGSAR SCTKVHKKGI 720 ALGRSVDLTK FSDYDELTAE LDQLFEFRGE LISPQKDWLV VFTDNEGDMM LVGDDPWQEF 780 CSMVRKIYIY PKEEIQKMSP GTLSSKNEEN HSATDGGDAQ ETKSQLNQSA SDN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-178 | 39 | 392 | 2 | 354 | Auxin response factor 1 |
| 4ldw_A | 1e-178 | 39 | 392 | 2 | 354 | Auxin response factor 1 |
| 4ldw_B | 1e-178 | 39 | 392 | 2 | 354 | Auxin response factor 1 |
| 4ldx_A | 1e-178 | 39 | 392 | 2 | 354 | Auxin response factor 1 |
| 4ldx_B | 1e-178 | 39 | 392 | 2 | 354 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Mtr.13210 | 0.0 | flower| glandular trichome| leaf| pod| root| stem | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the sepals and carpels of young flower buds. At stage 10 of flower development, expression in the carpels becomes restricted to the style. Also expressed in anthers and filaments. At stage 13, expressed in the region at the top of the pedicel, including the abscission zone. Expressed in developing siliques. {ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant. {ECO:0000269|PubMed:10476078, ECO:0000269|PubMed:15960614}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Could act as transcriptional activator or repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Functions independently of ethylene and cytokinin response pathways. May act as a repressor of cell division and organ growth. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952, ECO:0000269|PubMed:16339187}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
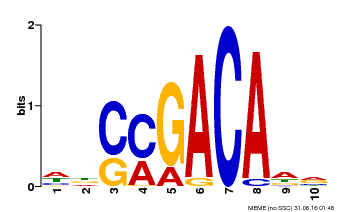
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Medtr8g100050.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC126794 | 0.0 | AC126794.15 Medicago truncatula chromosome 4 clone mth2-24j7, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003630583.2 | 0.0 | auxin response factor 2B | ||||
| Swissprot | Q94JM3 | 0.0 | ARFB_ARATH; Auxin response factor 2 | ||||
| TrEMBL | G7LIT1 | 0.0 | G7LIT1_MEDTR; Auxin response factor | ||||
| STRING | AET05059 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3281 | 33 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Medtr8g100050.1 |
| Entrez Gene | 11439423 |



