 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.B00609.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 968aa MW: 108543 Da PI: 6.5759 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 180.1 | 2.7e-56 | 20 | 136 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
+e ++rwl++ ei++iL n++k++++ +++++p sgs++L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg++++l+cyYah+e+n++f
Migut.B00609.2.p 20 SEaQHRWLRPAEIVEILRNYQKFHISPAAPNKPVSGSVFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDMLHCYYAHGEDNENF 112
4449***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+ywlLe++l +iv++hylevk
Migut.B00609.2.p 113 QRRSYWLLEQDLMHIVFAHYLEVK 136
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.378 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.0E-78 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.4E-49 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.2E-6 | 380 | 467 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 7.0E-5 | 381 | 466 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 5.13E-15 | 381 | 467 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 7.07E-14 | 579 | 672 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.3E-16 | 579 | 676 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.2E-16 | 580 | 674 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.271 | 580 | 674 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.1E-7 | 585 | 672 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 9.6E-5 | 613 | 642 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 10.659 | 613 | 645 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2200 | 652 | 681 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.73 | 788 | 810 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.59 | 789 | 818 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 7.97E-8 | 791 | 839 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.0013 | 791 | 809 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 3.8E-4 | 811 | 833 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.109 | 812 | 836 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.9E-5 | 814 | 833 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 968 aa Download sequence Send to blast |
MAESGSSSLG FRLDIKQILS EAQHRWLRPA EIVEILRNYQ KFHISPAAPN KPVSGSVFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDMLH CYYAHGEDNE NFQRRSYWLL 120 EQDLMHIVFA HYLEVKGNKT NISSSRNSDT VVSNSENNSV LLSSSFHGTS PSSTLSSACE 180 DAESEDNHQA SSRFHSFPES PLTDGSHYAQ SNSYNPTSSP GQTAFKQESG CYLPVQANWQ 240 GNTSFEEKSL HVNQENFVRP FYTLPGEQKE QSEQKNLQML LSEAETGHAM NQNMENLSSM 300 GNENYSLFLK KPSISGLQKE ENLKKADSFS RWIAKELEDY DELNLQSNNG ISWSGYGMPA 360 QLQVDLDTLN PSISQDQLFS IMDFSPNWAY TNMKTKVLIT GIFLKSEQEL SKCRWSIMFG 420 QVEVAAEVLA DGILCCHAPL QKPGLVPFYV TCSNRLACSE IREFEYRFEQ DQSMGGIDER 480 GSTNVMHLYQ RFETKLSLET SGSDLNSSGN DFGKQNIINK IFSFMEEENN QEIKLTPEKD 540 TTELMVIGEL LLQKQLKEKF YTWLFHKLSY DSESIADVKG GQGVLHLAAA LGFNWVLQPI 600 IVSGISIDFR DVNGWTALHW AAHYGREDTV AALVSLGASP GALTDPSAEY PLGRTPADLA 660 SSSGHKGISG FLAETTLTTH LSTLGVDDPL VHGGSGFSGS RAVQTVSERL AVPTTGEDVP 720 DTLSLKDSLA AICNATQAAA RIHQIFRTQS FHRKQLLEHG GDESVTPNEN AISLVAGKNS 780 RLGRASGGAN AAAVRIQKKF RGWKKRKEFL LIRQKVVKIQ AHFRGHQVRK KYKTIIWSVG 840 IMEKVILRWR RKRSGLRGFR SDAVAKVESG QGTLPQEDDY DFLKEGRKQT EERMQKALAR 900 VKSMAQYPEA RAQYRRLLTA AEGFRETKDA SDAEIQETSD DMFYPQDDLI DIESLLDDDT 960 FMSLAFQ* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
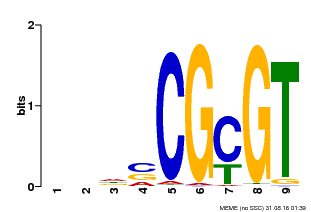
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.B00609.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012828340.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2 isoform X1 | ||||
| Refseq | XP_012828341.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2 isoform X1 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | A0A022PSL8 | 0.0 | A0A022PSL8_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.B00609.1.p | 0.0 | (Erythranthe guttata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.B00609.2.p |



