 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.B01828.3.p | ||||||||
| Common Name | MIMGU_mgv1a008458mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 374aa MW: 40312.8 Da PI: 6.9828 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 70.8 | 2.1e-22 | 245 | 307 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
++elkrerrkq+NRe+ArrsR+RK+ae+eeL+ kv++L++eN++Lk e+++l + ++ lk e+
Migut.B01828.3.p 245 DRELKRERRKQSNRESARRSRLRKQAETEELAVKVQALTTENMTLKAEINKLMESSEHLKLEN 307
689*********************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 3.0E-32 | 1 | 94 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 1.5E-12 | 115 | 232 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 9.1E-16 | 239 | 304 | No hit | No description |
| SMART | SM00338 | 3.2E-21 | 245 | 309 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 9.0E-21 | 246 | 307 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.886 | 247 | 310 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.13E-11 | 248 | 304 | No hit | No description |
| CDD | cd14702 | 7.18E-21 | 250 | 298 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 252 | 267 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 374 aa Download sequence Send to blast |
MGSNEEAKPS KAEKPSSPVT DQNNSVHVYP DWAAMQAFYG PRFAVPPYMN SGVASPHAPV 60 PYMWGPPQSM MPPYGPPYAA FYAHGGVYAH PGVPVASTAL SMETPVKSPG NTDGGFVKKL 120 KEFDGLAMSI GNGNGDSTEH DTDRRLSDSE ETDGSSDGSN GVTPRAGQNG KKRSRQGSPS 180 SGDGKANKKS NLVTAVEVGE KTTDVIPSAV KTMENTNTAL ELNDPSQNVK SSPTNAPQPL 240 MQNNDRELKR ERRKQSNRES ARRSRLRKQA ETEELAVKVQ ALTTENMTLK AEINKLMESS 300 EHLKLENTAL AEKLRGAQVE QTEEANLHKI DELRMKPVGT VNLLARVNNN SDGEPKLHQL 360 LDSNPRTDAV AAG* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 261 | 267 | RRSRLRK |
| 2 | 261 | 268 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the G-box-like motif (5'-ACGTGGC-3') of the chalcone synthase (CHS) gene promoter. G-box and G-box-like motifs are defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00318 | DAP | Transfer from AT2G46270 | Download |
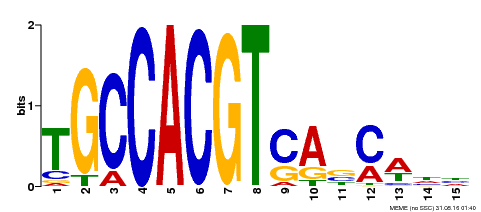
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.B01828.3.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012843813.1 | 0.0 | PREDICTED: common plant regulatory factor 1 isoform X3 | ||||
| Swissprot | Q99089 | 1e-122 | CPRF1_PETCR; Common plant regulatory factor 1 | ||||
| TrEMBL | A0A022R074 | 0.0 | A0A022R074_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.B01828.1.p | 0.0 | (Erythranthe guttata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46270.1 | 2e-89 | G-box binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.B01828.3.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



