 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.D01829.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 432aa MW: 45236.5 Da PI: 7.8123 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 136.5 | 2.3e-42 | 102 | 260 | 2 | 133 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssasec.eaesssssasnsssg..... 88
++k+++++++hTkv+gR+RR+R++a+caar+F+L++eLG+++d++tieWLlqqa+pa++++tgt++++a+++ + s +ss+s+ s +
Migut.D01829.1.p 102 PPKRTSTKDRHTKVDGRGRRIRMPALCAARVFQLTRELGHKSDGETIEWLLQQAEPAVIAATGTGTIPANFTsLNISLRSSGSSMSVPsqlrs 194
789******************************************************************999877777777775555577778 PP
TCP 89 .....................kaaksaakskksqksaasalnlakesrakarararertrekmrik 133
++++s+a+ + q+ a+ + ++a+++++++r+++ + t ++++ +
Migut.D01829.1.p 195 tyfnpnysmpqrrgffpgiglSSESSSATLLNFQAGANLNPSFAHNTKQEMRENSLDLTDQNQAAE 260
88999999999999*****99555555555555666666688888888888888777666555544 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 4.0E-44 | 106 | 277 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 27.923 | 108 | 162 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008283 | Biological Process | cell proliferation | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0010029 | Biological Process | regulation of seed germination | ||||
| GO:0010229 | Biological Process | inflorescence development | ||||
| GO:0031347 | Biological Process | regulation of defense response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 432 aa Download sequence Send to blast |
MEGGDDHLHL HHHNHHHHHH NQQQHHHHSR PNFPFQLLEK KEDHESTSSA AAAYPSLAIS 60 ADVASNPSTS TTPRSTATAS SSLQIAAVGG AAAASDTSKK PPPKRTSTKD RHTKVDGRGR 120 RIRMPALCAA RVFQLTRELG HKSDGETIEW LLQQAEPAVI AATGTGTIPA NFTSLNISLR 180 SSGSSMSVPS QLRSTYFNPN YSMPQRRGFF PGIGLSSESS SATLLNFQAG ANLNPSFAHN 240 TKQEMRENSL DLTDQNQAAE ESLGRKRRAA AAEQQELQPQ HHHHMGSYLL QSSTGAMPAS 300 HASIPANFWM VANSNNNQVM GGDPIWTFPS VNINNSSAAA AAALYRGTMS SGLHFMNFPA 360 PVALLPSGGG GGSGGGGGGS GNMGDGQLGM LAGLNTYRGG GGSGVSDSQA SGSHGGGGGG 420 DDRHDDNSHH S* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 366 | 379 | SGGGGGSGGGGGGS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved the regulation of plant development. Together with TCP15, modulates plant stature by promoting cell division in young internodes. Represses cell proliferation in leaf and floral tissues (PubMed:21668538). Together with TCP15, acts downstream of gibberellin (GA), and the stratification pathways that promote seed germination. Involved in the control of cell proliferation at the root apical meristem (RAM) by regulating the activity of CYCB1-1 (PubMed:25655823). Involved in the regulation of seed germination. May regulate the activation of embryonic growth potential during seed germination (PubMed:17953649, PubMed:22155632). Acts together with SPY to promote cytokinin responses that affect leaf shape and trichome development in flowers (PubMed:22267487). Transcription factor involved in the regulation of endoreduplication. Represses endoreduplication by activating the gene expression of the key cell-cycle regulators RBR1 and CYCA2-3 (PubMed:25757472). Regulates the expression of the defense gene pathogenesis-related protein 2 (PR2) in antagonism to SRFR1, a negative regulator of effector-triggered immunity (PubMed:24689742). Involved in positive regulation of plant defense. Represses jasmonate (JA) response to promote disease resistance. Regulates the plant immune system by transcriptionally repressing a subset of JA-responsive genes (PubMed:28132837). {ECO:0000269|PubMed:17953649, ECO:0000269|PubMed:21668538, ECO:0000269|PubMed:22155632, ECO:0000269|PubMed:22267487, ECO:0000269|PubMed:24689742, ECO:0000269|PubMed:25655823, ECO:0000269|PubMed:25757472, ECO:0000269|PubMed:28132837}. | |||||
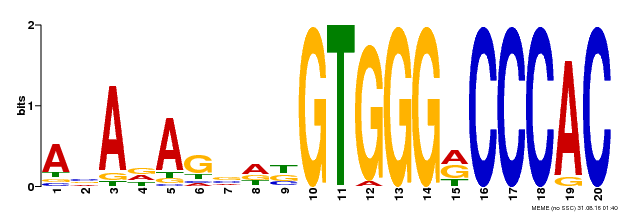
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00396 | DAP | Transfer from AT3G47620 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.D01829.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced during seed imbibition (PubMed:17953649). Circadian-regulation with the lowest expression in the middle of the dark period (PubMed:17953649). {ECO:0000269|PubMed:17953649}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012853009.1 | 0.0 | PREDICTED: transcription factor TCP14-like | ||||
| Swissprot | Q93Z00 | 4e-65 | TCP14_ARATH; Transcription factor TCP14 | ||||
| TrEMBL | A0A1S4A6U8 | 1e-170 | A0A1S4A6U8_TOBAC; transcription factor TCP14-like | ||||
| STRING | Migut.D01829.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2882 | 23 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47620.1 | 6e-49 | TEOSINTE BRANCHED, cycloidea and PCF (TCP) 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.D01829.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



