 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.E00229.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 541aa MW: 61838.6 Da PI: 5.9848 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 76.9 | 2.6e-24 | 283 | 336 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++Wt +LH++Fv+av+q+ G ek Pk+il+lm+v+ Lt+e+v+SHLQkYRl
Migut.E00229.2.p 283 KARVVWTVDLHQKFVRAVNQI-GFEKVGPKKILDLMGVPWLTRENVASHLQKYRL 336
68*******************.9*******************************8 PP
| |||||||
| 2 | Response_reg | 83.6 | 6e-28 | 102 | 210 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealk 91
vl+vdD+p+ +++l+++l+k +y ev+++ ++eal++l+e++ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + + ++
Migut.E00229.2.p 102 VLVVDDDPTWLKILEKMLKKCNY-EVTTCNLAREALNILRERKdgFDIVISDVNMPDMDGFRLLEHVGLEM-DLPVIMMSVDGETSRVMKGVQ 192
89*********************.***************999999**********************6644.8******************** PP
TTESEEEESS--HHHHHH CS
Response_reg 92 aGakdflsKpfdpeelvk 109
Ga d+l Kp+ ++el++
Migut.E00229.2.p 193 HGACDYLLKPIRMKELRN 210
***************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.40.50.2300 | 3.0E-44 | 99 | 223 | No hit | No description |
| SuperFamily | SSF52172 | 2.56E-36 | 99 | 223 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 1.9E-32 | 100 | 212 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.462 | 101 | 216 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 5.6E-25 | 102 | 211 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.26E-30 | 103 | 216 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-28 | 281 | 341 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.23E-17 | 281 | 340 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.4E-21 | 283 | 336 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 8.2E-8 | 285 | 335 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 541 aa Download sequence Send to blast |
MTVWCPHRPL TPSYIAGAKY SFILSLDLFP LKNPLSLSLF LSSTLFLSLV HTVCSIIYIY 60 EPYTYIIVCR CLFSFRLITN LMMESSGFSS PRTECFPAGL RVLVVDDDPT WLKILEKMLK 120 KCNYEVTTCN LAREALNILR ERKDGFDIVI SDVNMPDMDG FRLLEHVGLE MDLPVIMMSV 180 DGETSRVMKG VQHGACDYLL KPIRMKELRN IWQHVFRKRI HEVRDIEGYE SIDEINNMMR 240 CGTEQFEDGF LFSGDFITGK KRKDVDSKYD DRVCGESSSS AKKARVVWTV DLHQKFVRAV 300 NQIGFEKVGP KKILDLMGVP WLTRENVASH LQKYRLYLSR LQKENELKST FGATKELDIF 360 SKDHSISAHQ IIQANETYAD KICDKNDLAK TSASIRNDFY PDVKSEPKIQ VHYSWNSEAP 420 PLELIKKDGN NKTVHFHSEY TFDKRAQTGP TPWNTSGPHS FDVLETPSTA EAAIRLWDEE 480 FNDYSPQGDF YPPINAYIGY DNQELIDGVP PHLYDSLKFD SSVGPGESVE YHVIDQGLFA 540 * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 6e-18 | 279 | 341 | 1 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
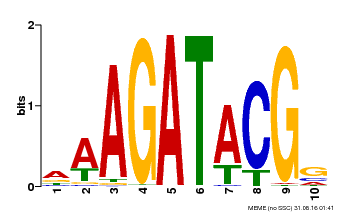
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.E00229.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012855343.1 | 0.0 | PREDICTED: two-component response regulator ARR11-like isoform X2 | ||||
| TrEMBL | A0A022Q1S7 | 0.0 | A0A022Q1S7_ERYGU; Uncharacterized protein | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-122 | response regulator 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.E00229.2.p |



