 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.E00311.1.p | ||||||||
| Common Name | LOC105966198, MIMGU_mgv1a008281mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 380aa MW: 41890.1 Da PI: 6.8331 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 97.5 | 9.3e-31 | 210 | 265 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k r++W+peLH+rF++ ++qLGGs+ AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Migut.E00311.1.p 210 KSRRCWSPELHRRFLQSLQQLGGSHLATPKQIRELMKVDGLTNDEVKSHLQKYRLH 265
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-27 | 207 | 268 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 13.088 | 207 | 267 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.03E-17 | 208 | 268 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.8E-26 | 210 | 265 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.6E-6 | 212 | 263 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 380 aa Download sequence Send to blast |
MSSINGHNHS DYSEKMQRCQ DYVHGLEQER SKIQVFQREL PLCLELVTQA IEACKQQLHG 60 QLSECSEQTS SDVVPVLEEF IPIKRAFSSF HSDEDDEEEE EQESKKLKND DDSSNKKSDW 120 LRSVQLWNNQ TPDLSKEDSP RKVAVTEVKR NGGGGAFHPF KKEKSTTTTT VAVVAAPPQE 180 GSMPPASTSS TAETGGGNKK EDKESQSQRK SRRCWSPELH RRFLQSLQQL GGSHLATPKQ 240 IRELMKVDGL TNDEVKSHLQ KYRLHTRRPN NQSMQNNNNN NQQAPQFVVV GGIWMQPEYA 300 TPMAAATTTS GEAASGVANS TGIYAPVASL PPPFHHASKQ RQHISDNDRG CSHSDGGVHS 360 NSPATSSSTH TSTASPAIY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-13 | 210 | 263 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate signaling in roots. {ECO:0000250|UniProtKB:Q9FX67}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00160 | DAP | Transfer from AT1G25550 | Download |
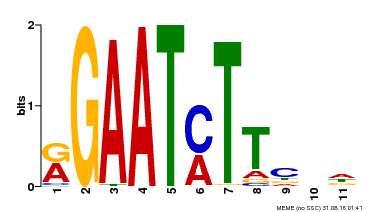
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.E00311.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012846219.1 | 0.0 | PREDICTED: uncharacterized protein LOC105966198 | ||||
| Swissprot | Q9FPE8 | 1e-100 | HHO3_ARATH; Transcription factor HHO3 | ||||
| TrEMBL | A0A022QMP6 | 0.0 | A0A022QMP6_ERYGU; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA5219 | 24 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68670.1 | 4e-76 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.E00311.1.p |
| Entrez Gene | 105966198 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



