 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.G00007.2.p | ||||||||
| Common Name | MIMGU_mgv1a000988mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 922aa MW: 102300 Da PI: 7.4723 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.2 | 1.6e-40 | 129 | 206 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C+adls+ak+yhrrhkvC+ hsk++++lv +++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Migut.G00007.2.p 129 VCQVEDCRADLSNAKDYHRRHKVCDLHSKSTSALVGDVMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTHP 206
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 6.1E-32 | 124 | 191 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.684 | 127 | 204 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.83E-36 | 129 | 207 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 7.4E-29 | 130 | 203 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 2.92E-4 | 674 | 808 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 2.0E-4 | 707 | 807 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 922 aa Download sequence Send to blast |
MESKIGGEKI HKSMEWDSNE WRWDGALFVA TPLNSVPSDC RSRQLGSDEL VLLGDGNGNG 60 REKRELEKRT RGVLEMEENE SEDVGLPNLK LGGQVYPIAE RDMYEMEGKS GKKTKVSSAL 120 PMSTSSRSVC QVEDCRADLS NAKDYHRRHK VCDLHSKSTS ALVGDVMQRF CQQCSRFHVL 180 QEFDEGKRSC RRRLAGHNKR RRKTHPENDE PANSSDQIQD QDLVSHLLKN LAHLTGTNPA 240 GVLPISQNVG TSLGTALKGL SAPSGPGVTI PASDLTEKRT LIGGVSHAST SESPLPFRTT 300 SSDLFKEKDS NTGVGRTKLS NIDLNCAYDG SQDCMEDMPN TSHLNKTSPG GSSWLCKDSQ 360 RCGPPQNSGN SASTSSQSPS TSSGEAQSRT DRIVFKLFGK DPSDFPLLLR KQILDWLSNS 420 PTDIESYIRP GCIILTIYLR MEKSSWDELY CNLTSSLLRL LNSSTDSFWR TGWIYTRVHH 480 HVTFMYNGQV VLDTPLPVRN HQSCRISSIK PIAVTVSEGV HFVVKGFNLS RSTSRLLCAL 540 EGKYLVQENC ADMTGRADSL TEHDQIQSLT FSCAVPNIVG RGFIEIEDHG LSSSFFPFIV 600 AEKDVCSEIC SLESVIEDAN EIQVRNEALD FIHEMGWLLQ RNRLKSRLGD GDLFPFERFR 660 RLTEFSIDHD WCAVVKKLLR ILFDDGTVDL GPHNSNIVAL LNDVGLVHRA VRRKCSSMVR 720 FLLNEKNPLA DGGGGAHLYL FRPDAAGPGG LTPLHIAASL DGCENVLDAL TEDPGSVGIE 780 EWKRGRDSSV LTAHDYACIR GQYSYINIVQ RKVDKKSTVV GVGVDIGDSS SSRGEVVLSV 840 SVDKKMEIER RRRRCGECEE RIMRYGNRST RGRVRIYRPA MLSLVGIAAV CVCTALLFKS 900 SPEVLFSFRP FRWDQLKYGS Q* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 7e-30 | 129 | 203 | 10 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 198 | 204 | KRRRKTH |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
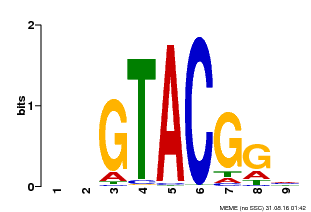
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.G00007.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT717966 | 1e-151 | KT717966.1 Erythranthe guttata squamosa promoter-binding protein 3 mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012842609.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 12 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | A0A022R2N5 | 0.0 | A0A022R2N5_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.G00007.1.p | 0.0 | (Erythranthe guttata) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.G00007.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



