 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.I00005.1.p | ||||||||
| Common Name | MIMGU_mgv1a009285mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 348aa MW: 38380.5 Da PI: 7.3347 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 124.4 | 3.6e-39 | 43 | 103 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
+e++ kcprC+st+tkfCyynnyslsqPryfCk+CrryWtkGG+lrn+PvGgg+rknkk+s
Migut.I00005.1.p 43 HEQSPKCPRCESTHTKFCYYNNYSLSQPRYFCKTCRRYWTKGGTLRNIPVGGGCRKNKKVS 103
67889*****************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 5.0E-27 | 31 | 97 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.1E-32 | 45 | 101 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.658 | 47 | 101 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 49 | 85 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010067 | Biological Process | procambium histogenesis | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 348 aa Download sequence Send to blast |
MDSSPISTPS GGDEENNINS ILACGGRSPT LMAAERSRLR PQHEQSPKCP RCESTHTKFC 60 YYNNYSLSQP RYFCKTCRRY WTKGGTLRNI PVGGGCRKNK KVSSSSSASA ANKKSDHHPP 120 PPPSSALVPL NRNNHNHNNN PSADDDENHV HLGFSDQLHF SSLHASGFFN NAGSNNNFVL 180 DNNNNNNIIN NHHTPIDFMD NKYGDFGGMM ITGSGGGGNN NADDHNAVNF HGFPFGLSSS 240 FLDHHHGSSN SAPINILESC QRLMLPYDNP NQNQNHNHDY NNHNHNHNNN HEVIDVKPNS 300 KILSLEWNDY HNHHHHQQST GYNFGGLGSW TGLMNGYGSN PATNPLV* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00580 | DAP | Transfer from AT5G62940 | Download |
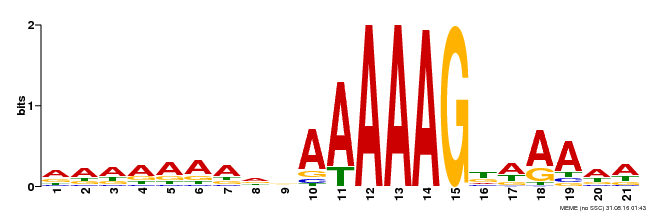
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.I00005.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF542507 | 5e-45 | AF542507.1 Oryza sativa Dof-like protein 34 mRNA, partial cds. | |||
| GenBank | AK376388 | 5e-45 | AK376388.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3122H18. | |||
| GenBank | AM084360 | 5e-45 | AM084360.1 Hordeum vulgare subsp. vulgare dof12 gene for dof zinc finger protein 12, exons 1-2. | |||
| GenBank | CP012610 | 5e-45 | CP012610.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 2 sequence. | |||
| GenBank | FP100350 | 5e-45 | FP100350.1 Phyllostachys edulis cDNA clone: bphylf036a01, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012831218.1 | 0.0 | PREDICTED: dof zinc finger protein DOF5.6 | ||||
| TrEMBL | A0A022RR81 | 0.0 | A0A022RR81_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.I00005.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4895 | 22 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62940.1 | 4e-35 | Dof family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.I00005.1.p |



