 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.I00266.1.p | ||||||||
| Common Name | MIMGU_mgv1a000334mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1223aa MW: 136944 Da PI: 8.5573 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.2 | 0.00026 | 1131 | 1153 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
Migut.I00266.1.p 1131 CPvkGCGKKFFSHKYLVQHRRVH 1153
9999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 12.2 | 0.00054 | 1189 | 1215 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
Migut.I00266.1.p 1189 YVCTetGCGQTFRFVSDFSRHKRKtgH 1215
99********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 9.9E-16 | 22 | 63 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.659 | 23 | 64 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 1.3E-14 | 24 | 57 | IPR003349 | JmjN domain |
| SuperFamily | SSF51197 | 7.14E-27 | 109 | 166 | No hit | No description |
| PROSITE profile | PS51184 | 33.03 | 187 | 356 | IPR003347 | JmjC domain |
| SMART | SM00558 | 1.9E-50 | 187 | 356 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 7.14E-27 | 205 | 367 | No hit | No description |
| Pfam | PF02373 | 3.0E-37 | 220 | 339 | IPR003347 | JmjC domain |
| SMART | SM00355 | 24 | 1106 | 1128 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.445 | 1129 | 1158 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0045 | 1129 | 1153 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 8.5E-6 | 1130 | 1152 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1131 | 1153 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.93E-9 | 1145 | 1187 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-9 | 1153 | 1181 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.679 | 1159 | 1188 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0018 | 1159 | 1183 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1161 | 1183 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.82E-8 | 1177 | 1212 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.0E-8 | 1182 | 1212 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.572 | 1189 | 1220 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.19 | 1189 | 1215 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1191 | 1215 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1223 aa Download sequence Send to blast |
MAAEVGGGGS IEVSPWLKSM PVAPEYHPTL AEFQDPIAYI FKIEKEASKY GICKIVPPVP 60 AAPRKTIVAN LNKSLLARSN NPDPGPTFTT RQQQIGFCPR KHRPVQKPVW QSGEKYTLAE 120 FETKAKNFEK NYLKKYSKKI LNPLEIETLY WNAMVDKPFQ VEYANDMPGS AFVPQKSGVK 180 KNESALTVGD TEWNMRRVSR ENLSLLRFMK EEIPGVTSPM VYVAMMFSWF AWHVEDHDLH 240 SMNFLHMGAG KTWYGVPREA AVAFEEVIRE HGYSGEINPL VTFATLGEKT TVMPPEVLLS 300 AGVPCCRLVQ NAGEFVVTFP RAYHSGFSHG FNCGEAANIA TPEWLRVARE AAIRRAAINC 360 PPMVSHFQLL YDLALSLCSR APKSIAAEPR SSRLKDRKKG EGEMLIKELF FQDMMQNNDM 420 LHILGKRSPI VLLSKNSLDS PSGSHSAAKS RLFPSLCSPD LEMKTTSNNN NAPDELICMK 480 QTKGHFRNSE EVPCMDREIK KACQKSEQGL FSCVTCGILC FACVAIVQPT EASARYIMSG 540 DCSIFNFWET SDNEHNDIKD AKAPNAKLSS SALMIGKTHS GRVFDAPLSV EKENSVGVVS 600 EKANKAPSSL GLLALTYANS SDSEEEDENE ADISFQGGGN CKIDSPENDT DLRMSDSNTK 660 FGLPIETHGN GESRNLTNNC NVAESKNSLT DRFRRQMESW NETSNSLTRK TEANNGSTPL 720 AESTMPFSSR SDEDSSRLHV FCLQHAMQVE KRLGEVGGAH VFLICHPDYP KLESQARKIA 780 EELENDSPWN EISFQDATEA DEEIIRLSLE SENSIHGNRD WAVKLGINLF YSANLSRSPL 840 YCKQMHYNSV IYGAFGRSSE IDDTSSIKAE IEGKSLGFGR HKKIFVAGKW CGKVWMSSHA 900 HPLLVDHDFL QEPDFKNERQ SSQRKRKSSV AENSAETTTK MDESSLDFVL RNCRKQIKRK 960 RGSRRMKEEN HEPEISDDSS EECRTKQLKK ETAVNLDDDS SDEFPLSSSW KQIKNKRGAN 1020 QEPVKSQPKT KKQIDEPEGG PSTRLRKRTK TLICKETGPS KAKPAPKKQQ NDAVIPAKAA 1080 KAKSPAAIKN PAKAKNQNRG DEEAEYLCDM EGCAMSFASK NELTLHKRNI CPVKGCGKKF 1140 FSHKYLVQHR RVHMDDRPLK CPWKGCKMTF KWAWARTEHV RVHTGARPYV CTETGCGQTF 1200 RFVSDFSRHK RKTGHTPKKA RG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 3e-74 | 16 | 377 | 8 | 353 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 3e-74 | 16 | 377 | 8 | 353 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 922 | 927 | QRKRKS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
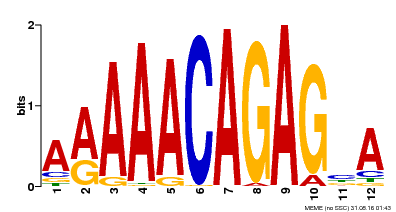
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.I00266.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012834201.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A022RIV6 | 0.0 | A0A022RIV6_ERYGU; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4988 | 23 | 31 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.I00266.1.p |



