 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.K00222.1.p | ||||||||
| Common Name | LOC105976534, MIMGU_mgv1a011578mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 278aa MW: 31487.3 Da PI: 6.5094 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.9 | 3.6e-16 | 15 | 62 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd lv +++++G+g+W +++ g+ R+ k+c++rw +yl
Migut.K00222.1.p 15 KGPWTPEEDIVLVSYIQEHGPGNWFAVPTNTGLLRCSKSCRLRWTNYL 62
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 45.2 | 2.1e-14 | 68 | 113 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+ E+ ++++ +++G++ W++Ia++++ Rt++++k++w+++l
Migut.K00222.1.p 68 RGNFTPHEEGMIIHLQALFGNK-WAAIASYLP-QRTDNDIKNYWNTHL 113
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 23.139 | 10 | 66 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.24E-29 | 13 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-13 | 14 | 64 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.0E-15 | 15 | 62 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-21 | 16 | 69 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.02E-10 | 17 | 62 | No hit | No description |
| SMART | SM00717 | 2.0E-13 | 67 | 115 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.638 | 67 | 117 | IPR017930 | Myb domain |
| Pfam | PF00249 | 6.4E-13 | 68 | 113 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.05E-10 | 70 | 113 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-25 | 70 | 118 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 278 aa Download sequence Send to blast |
MGRPPPSCDK VVIKKGPWTP EEDIVLVSYI QEHGPGNWFA VPTNTGLLRC SKSCRLRWTN 60 YLKPGIKRGN FTPHEEGMII HLQALFGNKW AAIASYLPQR TDNDIKNYWN THLKKKLKKI 120 QAQSPPLTPH VAYDSTKPRN HSDLKITENS SVYASSSQNI SSLLQEWNNT STTCDLDRNI 180 RNIGKIPTEF SDDYSTLLQY YRPQQQQFDC SILSKDRSFI EEKSSCDSSQ KGSDQSSGPI 240 TDQEKIDDNP APLSFLDEWL LDASASQVED MEIPQMF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 5e-24 | 13 | 117 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the regulation of gene (e.g. drought-regulated and flavonoid biosynthetic genes) expression and stomatal movements leading to negative regulation of responses to drought and responses to other physiological stimuli (e.g. light). {ECO:0000269|PubMed:22018045}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00132 | DAP | Transfer from AT1G08810 | Download |
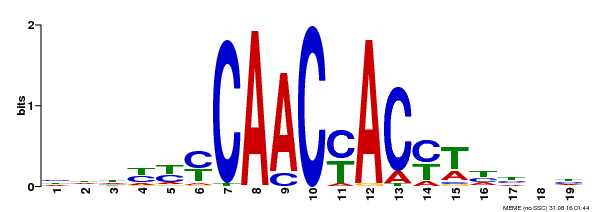
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.K00222.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by abscisic acid (ABA) and osmotic stress (salt stress). {ECO:0000269|PubMed:22018045}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012857223.1 | 0.0 | PREDICTED: myb-related protein 306-like | ||||
| Swissprot | B3VTV7 | 5e-96 | MYB60_VITVI; Transcription factor MYB60 | ||||
| TrEMBL | A0A022PZG2 | 0.0 | A0A022PZG2_ERYGU; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA12 | 24 | 2154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08810.1 | 3e-88 | myb domain protein 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.K00222.1.p |
| Entrez Gene | 105976534 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



