 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.N00052.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 918aa MW: 103845 Da PI: 7.2489 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 164.2 | 2.2e-51 | 26 | 143 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenpt 93
++e k+rwl+++ei+aiL n++ ++++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+ pt
Migut.N00052.1.p 26 MEEaKARWLRPNEIHAILYNHKCFTVHVKPMNLPKSGAILLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNSERIHVYYAHGEHSPT 118
45669**************************************************************************************** PP
CG-1 94 fqrrcywlLeeelekivlvhylevk 118
f rrcywlL+++le+ivlvhy+e++
Migut.N00052.1.p 119 FVRRCYWLLDKSLEHIVLVHYRETQ 143
**********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 74.51 | 22 | 148 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.0E-74 | 25 | 143 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 9.9E-46 | 28 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 2.8E-10 | 371 | 456 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 1.3E-16 | 553 | 666 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.83E-15 | 553 | 663 | No hit | No description |
| Pfam | PF12796 | 1.3E-7 | 553 | 633 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.87E-16 | 553 | 667 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.326 | 562 | 675 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 9.1E-6 | 604 | 633 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.915 | 604 | 636 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2000 | 643 | 674 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.72E-5 | 750 | 815 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.035 | 752 | 778 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 72 | 767 | 789 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.272 | 768 | 797 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0054 | 790 | 812 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.048 | 791 | 815 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 5.8E-5 | 792 | 811 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 15 | 871 | 893 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 3.72E-5 | 871 | 897 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 8.279 | 873 | 901 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 918 aa Download sequence Send to blast |
MESNRLVGSE IHGFHTMEDL DFVNMMEEAK ARWLRPNEIH AILYNHKCFT VHVKPMNLPK 60 SGAILLFDRK MLRNFRKDGH NWKKKKDGKT VKEAHEHLKV GNSERIHVYY AHGEHSPTFV 120 RRCYWLLDKS LEHIVLVHYR ETQELQGSPT TPGNSNSSSV ASDPSASWPL LEKSDSTVDR 180 VYEGDKRSLL ERDNSTTVEN HEQRLHEINT LEWDELLVPE QQENAASSEL TNLYLTCSFK 240 TMDDALSTNK ASPESSGDSF SGHVSGSTSI DYNVARNIPY QTAAHETIMN LQSFNSGLGP 300 LGGRNSIENP AKDHFKKLDS FEKWMTDIIA DSPGSVDNQT LESSFSTEHQ SFKSSTMDNH 360 LLSAVDQIFS ITDVSPSWAL STEETKILVV GFFNGQLPDT DFHLYLACGD SVVPVEVVQA 420 GVFRMVIPAQ TPGLVNLYLT FDGHKPISQV WPFEFRAPVV PHKTISSDDK PNWEEFQLQM 480 RLAHLLFSSD SLNIFSNKVS QNALKEAKIF AQRTAHIPNG WVHLTKLIQD AKVPFPQAKD 540 SLFELTLQNR LQEWLLEKVV SGCKIPERDE QGQGVIHLCA ILGYTWAVLP FSLSGLSMDY 600 RDKSGWTALH WAAYQGREKM VAALLSAGAK PNLVTDPTSA HPGGCTAADV ASKNGFDGLA 660 AYLAEKALVA QFNDMTLAGN VSGSLQITSN ETMDPGNFTE DELYLKDTLA AYRTAADAAA 720 RIHSAFREHS LSVRTRAVEA SNPEMEARNI VAAMKIQHAF RNYETRKQMA AAARIQYRFR 780 TWKMRRNFIN MRRHAIKIQA VFRGFQVRKH YCKILWSVGV VEKAILRWRK KRKGFRGLQV 840 QKSDEQDTPT PKDPNVEEDF FLASRKQAED RVERSVIRVQ AMFRSKQAQE DYRRMKLEHN 900 KATLEYEELL HPDVNMG* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
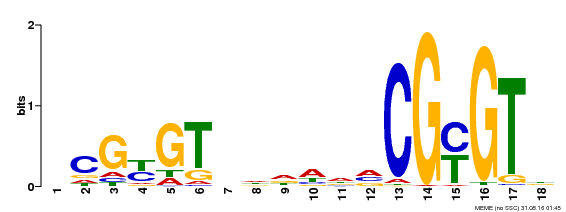
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.N00052.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012840434.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A2G9GJ79 | 0.0 | A0A2G9GJ79_9LAMI; Uncharacterized protein | ||||
| STRING | Migut.N00052.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4565 | 22 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.N00052.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



