 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Migut.N02994.1.p | ||||||||
| Common Name | LOC105959442, MIMGU_mgv1a010420mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Phrymaceae; Erythranthe
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 314aa MW: 35389.3 Da PI: 4.2912 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 27.9 | 5.1e-09 | 153 | 195 | 4 | 46 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLk 46
++ +r ++NR AA rsR+RKk++++ Le k+k eae ++L
Migut.N02994.1.p 153 ERKRKRQIRNRDAAVRSRERKKMYVKDLEMKSKYYEAECRRLG 195
57789999**************************999988775 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.9E-12 | 145 | 211 | No hit | No description |
| SMART | SM00338 | 8.1E-7 | 147 | 214 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF07716 | 4.2E-9 | 152 | 202 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.622 | 152 | 194 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.31E-10 | 154 | 210 | No hit | No description |
| CDD | cd14704 | 1.80E-13 | 155 | 206 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 157 | 172 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 314 aa Download sequence Send to blast |
MFDHEFVESE FIDWGQLLEN IPDNPNFDFC DPPPADLFSN SPDSGASLSV DDIEQYLMND 60 DSHPDEKYED NLVDEFLSDV LLDSSPGSGS DRSKDSSTSP NSVEVGTDRE EKEEEGDKSD 120 HSGADVPQTK ENGVIDAVDD EDDEDDEDDD PSERKRKRQI RNRDAAVRSR ERKKMYVKDL 180 EMKSKYYEAE CRRLGNFLQC CLAENQALRL SLHTNKAFDA SMSKQESAVL LLESLLLGSL 240 LGFLGIIYLL TLPSQLLSIL EKALLPNVGS AEWENVTPRK EASKVFRVQF HSFMMTKRCK 300 ASRSRMKLSP VVL* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 154 | 173 | KRKRQIRNRDAAVRSRERKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
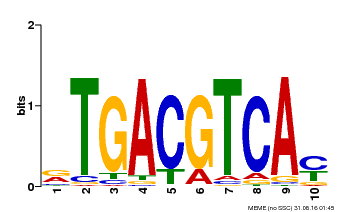
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Migut.N02994.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012838999.1 | 0.0 | PREDICTED: bZIP transcription factor 60-like | ||||
| TrEMBL | A0A022R8A0 | 0.0 | A0A022R8A0_ERYGU; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA7101 | 21 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 2e-16 | basic region/leucine zipper motif 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Migut.N02994.1.p |
| Entrez Gene | 105959442 |



