 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_000160-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 610aa MW: 66855.9 Da PI: 5.3616 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 463 | 1.7e-141 | 234 | 597 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalklfsevsPilkfshl 96
lv++L++cAeav++++l++a+al++++ la +++ +m+++a+yf+eALa+r+++ l p+++ + ss + + + f+e++P+lkf+h+
NNU_000160-RA 234 LVHTLMACAEAVQQNNLKMAEALVKQIGLLAVSQAGAMRKVATYFAEALARRIYQ--------LYPQDSLD--SSFSDMLQMHFYETCPYLKFAHF 319
689***************************************************9........55565554..4444445556************* PP
GRAS 97 taNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvakrledlele 192
taNqaIlea++g++rvH+iDf+++qG+QWpaL+qaLa Rp+gpp++R+Tg+g+p+++++++l+++g++La++A++++v+fe++ +va++l+dl+ +
NNU_000160-RA 320 TANQAILEAFAGKSRVHVIDFSMKQGMQWPALMQALALRPGGPPTFRLTGIGPPQQDNTDALQQVGWKLAQLAKSIHVKFEYRGFVANSLADLDAS 415
************************************************************************************************ PP
GRAS 193 eLrvkp..gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsl.eaklpreseerikv 285
+L+++p +Ea+aVn+v++lhrll++++++e+ vL+ +k+++Pk+v++veqea+hn++ Fl+rf+eal+yys+lfdsl +++ +++++++
NNU_000160-RA 416 MLDLRPseDEAVAVNSVFELHRLLARPGAIEK----VLSSIKEMKPKIVTIVEQEANHNGPVFLDRFTEALHYYSTLFDSLeGCAMSPPNSQDQVM 507
*****999************************....*********************************************777899999****** PP
GRAS 286 ErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvlgWkdrpLvsvSaWr 374
++ +lgr+i+nvvaceg +r+erhe l +Wr+rl+++GF+pv+l+ +a kqa++ll+ ++ +dgyrvee++g+l+lgW++rpL+++SaW+
NNU_000160-RA 508 SELYLGRQICNVVACEGVDRVERHEALTQWRNRLGSSGFSPVHLGFNAFKQASMLLALFAgGDGYRVEENNGCLMLGWHTRPLIATSAWQ 597
*****************************************************************************************6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01129 | 1.1E-37 | 37 | 110 | No hit | No description |
| Pfam | PF12041 | 2.1E-36 | 37 | 104 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| PROSITE profile | PS50985 | 67.981 | 208 | 576 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 5.9E-139 | 234 | 597 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 610 aa Download sequence Send to blast |
MKRELQESTS GGGVYSMAEA GKCKMWDEAE QQDAGVDELL AVLGYKVRSS DMAEVAQKLE 60 QLDIVMGSAQ EDGLNHLASE TVHYNPSDLA TWLESMLSEL NAPPIFDPTP SQPSLPRVLD 120 DPRLAPAESS SMVTIDFPNS RQQQQQPKQQ IHPQQPRIYD EPTDYDLRAI PGGAICGQVE 180 PASKRMKPTS TSTSASSSSS PSAAPAAVGV TSGVSTELAR PVVLVDSQET GVRLVHTLMA 240 CAEAVQQNNL KMAEALVKQI GLLAVSQAGA MRKVATYFAE ALARRIYQLY PQDSLDSSFS 300 DMLQMHFYET CPYLKFAHFT ANQAILEAFA GKSRVHVIDF SMKQGMQWPA LMQALALRPG 360 GPPTFRLTGI GPPQQDNTDA LQQVGWKLAQ LAKSIHVKFE YRGFVANSLA DLDASMLDLR 420 PSEDEAVAVN SVFELHRLLA RPGAIEKVLS SIKEMKPKIV TIVEQEANHN GPVFLDRFTE 480 ALHYYSTLFD SLEGCAMSPP NSQDQVMSEL YLGRQICNVV ACEGVDRVER HEALTQWRNR 540 LGSSGFSPVH LGFNAFKQAS MLLALFAGGD GYRVEENNGC LMLGWHTRPL IATSAWQLAS 600 DRHLRLAACL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3g_A | 2e-67 | 229 | 596 | 14 | 378 | Protein SCARECROW |
| 5b3h_A | 2e-67 | 229 | 596 | 13 | 377 | Protein SCARECROW |
| 5b3h_D | 2e-67 | 229 | 596 | 13 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. | |||||
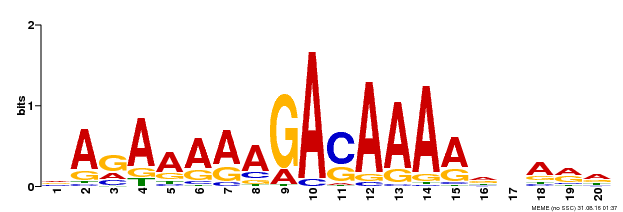
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010264458.1 | 0.0 | PREDICTED: DELLA protein GAI1 | ||||
| Swissprot | Q8S4W7 | 0.0 | GAI1_VITVI; DELLA protein GAI1 | ||||
| TrEMBL | A0A1U8APB6 | 0.0 | A0A1U8APB6_NELNU; DELLA protein GAI1 | ||||
| STRING | XP_010264458.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01570.1 | 0.0 | GRAS family protein | ||||



