 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_000795-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1085aa MW: 120066 Da PI: 8.6163 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 132.1 | 1.9e-41 | 155 | 232 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C+ dls+ak+yhrrhkvCe+hsk++++lv g++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+q+
NNU_000795-RA 155 MCQVDDCKGDLSNAKDYHRRHKVCEFHSKTTKALVGGQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQP 232
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.2E-35 | 149 | 217 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.359 | 153 | 230 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.57E-38 | 154 | 233 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.0E-30 | 156 | 229 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 3.1E-4 | 874 | 977 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.2E-6 | 874 | 970 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.06E-4 | 878 | 973 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1085 aa Download sequence Send to blast |
MEDVGAQVFP PIFIHQALPG RFCEAPAMAK KRDLPWQSPN FQQQQQQHQR FNSVFQSSKG 60 NWNPSSWDWD SMMFVAKPSE TEVLHLGTAA VVESEQKKKG EETLKNLVVK KGSVDEDGEK 120 LTLKLGGGLY SVDESAARPN KRVRSGSPGS GNYPMCQVDD CKGDLSNAKD YHRRHKVCEF 180 HSKTTKALVG GQMQRFCQQC SRFHPLSEFD EGKRSCRRRL AGHNRRRRKT QPEDASSRLL 240 GPGNRENSGT GNMDVVNLLT ILSRLQGNNV DRSANASSLP DRERLIQILN KINASPISGN 300 SGPRLPVPGS FDLNVSQEAS SDNLNKINGN TSSPSTMDLL AVLSAALAAS NPDALALLSQ 360 INNHSYDGDK SKLNSLDQAD GSRLQNKPIS RYTSIGGERN NGTFQSPVGT PDCHAQIPKS 420 SLPFQLFSSS PEGGSPPKLG SSRRYFSSES SNPMEERSPS SSPVVQKLFP LHAETEILKH 480 ERMSISGEDN ATVETSTTRD WTSPLELFKG QNGRVGNGSV QSLQYQGGYT SSSGSDHSPS 540 SSNSDAQDRT GRIIFKLFDK DPSNFPGTLR SQILNWLSHS PSEMESYIRP GCVVLSVYVS 600 MPSTAWEHFQ KNLFHLAKLL VQGSASDFWR NGRFLVHTDR QLVSHKDGKI RLCKAWRTWS 660 APELISVSPL AVVGGEETSL VLKGRNLTAP GTKIHCTYLG GYKTKEVPGS TYQVAMYDDT 720 SFERFKFPGG APGVLGRCFI EVENGFKGNC FPVIIADATI CQELRGLESE FDQVSRTACI 780 VTENKFQDLG RPQSREDVLH FLNELGWLFQ RKSNPSKPEG PNFSHSRFKF IFTFSVERDW 840 CAVVKTLLDI LVEKNLGPDG PPKASIELLS DIQLLNRAVK RKCRNMVDLL IHYSVTLGDN 900 TKQYLFPPNS VGPGGVTPLH LAACIQGLEE IVDSLTNDPQ QIGLKCWNSL PDANGQTPFT 960 YSLMRNNHSY NRMVARKLAE RKRGQVSIPV GDEISLDQSW IIDEQADKPL PETLQGRRSC 1020 ARCAVVATRY YKRMPGSQGL LHRPYVHSML AIAAVCVCVC LFLRGSPDIG SVAPFKWENL 1080 DYGWS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-32 | 146 | 229 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
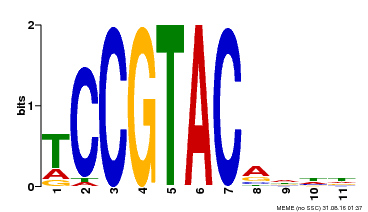
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010256977.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A1U7ZXH0 | 0.0 | A0A1U7ZXH0_NELNU; squamosa promoter-binding-like protein 14 | ||||
| STRING | XP_010256977.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



