 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_001186-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 365aa MW: 40720.8 Da PI: 9.4062 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 44.9 | 2.9e-14 | 60 | 108 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f +eeAa+a++ a+++++g
NNU_001186-RA 60 SQYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEEEAARAYDIAAQRFRG 108
78****9888.8*********3.....5*************************98 PP
| |||||||
| 2 | B3 | 101.9 | 3.6e-32 | 192 | 291 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE.. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefel 92
f+k+ tpsdv+k++rlv+pk++ae+h g++ +++ l +ed g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F+++ +++l
NNU_001186-RA 192 FDKAVTPSDVGKLNRLVIPKQHAEKHfplqIGSTSKGVLLNFEDNGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDVVSFQRSTGPDKQL 287
89***************************95566799**************************************************876677777 PP
EEEE CS
B3 93 vvkv 96
++ +
NNU_001186-RA 288 FIDW 291
7766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 4.32E-26 | 60 | 115 | No hit | No description |
| SuperFamily | SSF54171 | 9.81E-18 | 60 | 116 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 5.6E-9 | 61 | 108 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.2E-30 | 61 | 122 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.521 | 61 | 116 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.7E-21 | 61 | 116 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.5E-5 | 62 | 73 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.5E-5 | 82 | 98 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 7.1E-42 | 182 | 299 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 4.84E-30 | 190 | 292 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.36E-27 | 191 | 280 | No hit | No description |
| Pfam | PF02362 | 8.6E-29 | 192 | 291 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.6E-24 | 192 | 295 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.41 | 192 | 294 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 365 aa Download sequence Send to blast |
MDGSCIDESS SNSISVSPAP VSSLPEKKET ERLCRMGSGA SVVIDSDSGV EAESRKLPSS 60 QYKGVVPQPN GRWGAQIYEK HQRVWLGTFN EEEEAARAYD IAAQRFRGRD AVTNFKPLSE 120 TDEDDIETIF LNSHSKAEIV DMLRKHTYND ELEQSKRNYG RNGDVSKRNR GGGCFGDSGG 180 SDRAAKAREL LFDKAVTPSD VGKLNRLVIP KQHAEKHFPL QIGSTSKGVL LNFEDNGGKV 240 WRFRYSYWNS SQSYVLTKGW SRFVKEKNLK AGDVVSFQRS TGPDKQLFID WKPRAASTYM 300 ALPQLVHPLN PAAQPVEVVR LFGVNIFKIP VSSVVNSWNG KRLRDVDEFS SLEPCSKKQR 360 VLGAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 4e-56 | 189 | 297 | 11 | 120 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds specifically to bipartite recognition sequences composed of two unrelated motifs, 5'-CAACA-3' and 5'-CACCTG-3'. May function as negative regulator of plant growth and development. {ECO:0000269|PubMed:15040885, ECO:0000269|PubMed:9862967}. | |||||
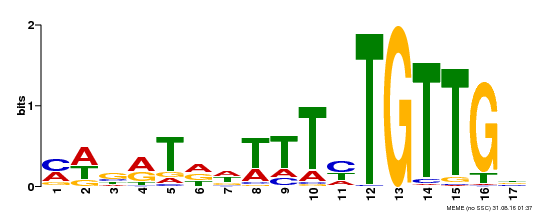
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by brassinosteroid and zeatin. {ECO:0000269|PubMed:15040885}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010262942.1 | 0.0 | PREDICTED: AP2/ERF and B3 domain-containing transcription factor RAV1-like | ||||
| Swissprot | Q9ZWM9 | 1e-148 | RAV1_ARATH; AP2/ERF and B3 domain-containing transcription factor RAV1 | ||||
| TrEMBL | A0A1U8AK51 | 0.0 | A0A1U8AK51_NELNU; AP2/ERF and B3 domain-containing transcription factor RAV1-like | ||||
| STRING | XP_010262942.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13260.1 | 1e-144 | related to ABI3/VP1 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



