 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_001809-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 334aa MW: 37113.8 Da PI: 4.6933 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59.4 | 5.9e-19 | 58 | 111 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ +q+++Le+ Fe ++++ e++ +LA++lgL+ rqV vWFqNrRa++k
NNU_001809-RA 58 KKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWK 111
566899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.7 | 2.4e-41 | 57 | 149 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekkrrls +qvk+LE++Fe e+kLeperKv+la+eLglqprqvavWFqnrRAR+ktkqlE+dy+ Lk++y++lke++++L++++e+L +e++e
NNU_001809-RA 57 EKKRRLSVDQVKALEKNFEVENKLEPERKVKLAQELGLQPRQVAVWFQNRRARWKTKQLERDYNLLKSNYETLKESHNALQQDKEALLAEIRE 149
69**************************************************************************************99986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.42E-19 | 43 | 115 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.508 | 53 | 113 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.3E-18 | 56 | 117 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 3.8E-16 | 58 | 111 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.60E-16 | 58 | 114 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 9.2E-20 | 60 | 120 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 9.7E-6 | 84 | 93 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 88 | 111 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 9.7E-6 | 93 | 109 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.3E-14 | 113 | 154 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 334 aa Download sequence Send to blast |
MKRLGSSESL GAVISICPSA EEKGLGASNH VYRRSFQSML DGLDEEDCLE EQGQISEKKR 60 RLSVDQVKAL EKNFEVENKL EPERKVKLAQ ELGLQPRQVA VWFQNRRARW KTKQLERDYN 120 LLKSNYETLK ESHNALQQDK EALLAEIREL KSKLGEENTE SNISVKQEAV MSESDNVATG 180 QSKIPAAGVL GGSEATEPNQ EGYKNSNANN GVGASGFTDF KDGSSDSDSS AILNEDNSPN 240 AAISSPGVFQ QNQILVSPTS SSLRFNCSSS SSPSSINCFQ FADSRGVSAS AQKAYQHQLM 300 RMEEHNFFNA EEPCNFFSED QAPSLQWYYP DQWN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 105 | 113 | RRARWKTKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
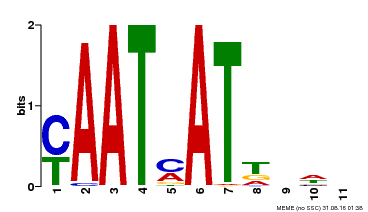
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010266919.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-6-like | ||||
| TrEMBL | A0A1U8AVF9 | 0.0 | A0A1U8AVF9_NELNU; homeobox-leucine zipper protein ATHB-6-like | ||||
| STRING | XP_010266919.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 1e-62 | homeobox protein 6 | ||||



