 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_004202-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1390aa MW: 155491 Da PI: 8.3343 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.9 | 0.00068 | 1274 | 1299 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++ Fs+k +L+ H r+ +
NNU_004202-RA 1274 YQCDieGCSMGFSSKQELVLHKRNiC 1299
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 11.2 | 0.0011 | 1299 | 1321 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
C+ Cgk F ++ +L++H r+H
NNU_004202-RA 1299 CSvkGCGKKFFSHKYLVQHRRVH 1321
77779****************99 PP
| |||||||
| 3 | zf-C2H2 | 11.9 | 0.00069 | 1357 | 1383 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
NNU_004202-RA 1357 YVCRepGCGQTFRFVSDFSRHKRKtgH 1383
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 4.3E-18 | 34 | 75 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.841 | 35 | 76 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 1.6E-13 | 36 | 69 | IPR003349 | JmjN domain |
| PROSITE profile | PS51184 | 34.37 | 202 | 371 | IPR003347 | JmjC domain |
| SMART | SM00558 | 8.3E-52 | 202 | 371 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.87E-26 | 216 | 375 | No hit | No description |
| Pfam | PF02373 | 8.3E-37 | 235 | 354 | IPR003347 | JmjC domain |
| SMART | SM00355 | 7.6 | 1274 | 1296 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.015 | 1297 | 1321 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.57 | 1297 | 1326 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.9E-5 | 1298 | 1325 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1299 | 1321 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.68E-9 | 1313 | 1355 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.2E-8 | 1326 | 1351 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0014 | 1327 | 1351 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.741 | 1327 | 1356 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1329 | 1351 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.83E-8 | 1345 | 1379 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.4E-9 | 1352 | 1380 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.845 | 1357 | 1388 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.25 | 1357 | 1383 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1359 | 1383 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1390 aa Download sequence Send to blast |
MRGFKCSSDL QLMAVVAAAE SPPEIFPWLK NLPLAPEYHP SVAEFQDPIA YILKIEKEAS 60 KFGICKIVPP LPPLPKKTVI ANINRSLAAR NNNSNSKSLP AFTTRQQQVG FCPRKSRPVQ 120 KPVWQSGETY TLQEFEAKAK QFEKTHLKKT GKKAISALEI ETLFWKASMD KPFSVEYAND 180 MPGSAFEPVN GKKWQEAGEA GSVGETAWNM RGVSRAKGSL LRFMKEEIPG VTSPMVYIGM 240 LFSWFAWHVE DHDLHSLNYL HMGAGKTWYG VPRDAAFAFE EVVRVHGYGE EVNPLVTFAI 300 LGEKTTVMSP EVLISTGIPC CRLVQNAGEF VVTFPRAYHS GFSHGFNCGE AANIATPEWL 360 RVAKEAAIRR ASINYPPMVS HFQLLYSLAL ALCSRIPMSI STEPRSSRLK DKRRGEGETM 420 VKELFVQNVV QNNDLIHVLL KKGSSCILLP HNSLDLSICS NLRVGSQRKV NPRLSLGLHS 480 PKEEMEASKI LLSDDMVLDR NTHLRNLSGF SSVKRKTSSV YERKSVPTLC GADYFCTSTT 540 EMHNLQTERV YNAGDGLLDQ GLFSCVVCGI LSFACTAIIQ PSEAAARYLV SADCSFFNDW 600 IVGSGVATDR YTVLDGDAGT AKLNSCSGME KCVRDGLYDV PVQSGDYQFQ VAGQSVEVTS 660 DTKTKTGISS LDLLAFAYGN SSDSEEDQDK PELSVFSDED DLKDSSSQCR STHSVLSSEH 720 TSLCNSSKLS LVHASTHKDG APSTLSCPSS TVSYASEVSV QVYASDSTPG HPSTNLKNQH 780 KKNFDTSLKS DTRSLVTLES NDLDDTYRDP LVVSRRVASK FTEVHHCDGK MDNIESERMK 840 SNDAETSVKS KAISLMQRTD NDSSRLHIFC LEHAVEVEKQ LNPIGGVHVL LLCHPEYLKI 900 EAEAKSLAEE LGIDYLWKDV PVREATEEDQ QRIQSALDDD EVIPSNGDWA VKLGINLYYS 960 ATISRSPLYS KQMPYNSVIY KAFGRSSPDI SPTRSKDSGK RPGKQKKIVV AGRWCGKVWM 1020 SNQVHPYLAQ ADESEEQELT RSFPIQTMPN RKPDRKVDIS QQEQPVLPEK IPLRGTMTTV 1080 AKNPGRKRKR SEKGMIKKQN CLQRDNPITS IDDSPELDTP SPCGRLIRSE QMKHETPPHH 1140 NDCEDIREED SCEKDDIEGG LSMRLRKRPQ KKPYEEVKVK PMHEKQTKKK SKKTSSSNEV 1200 DTKDEEADYL CNIEGCSCKK DGIEGGLSMR LRKRPQKKPC EVKVKPMNEK QMKKKGKETS 1260 SSNEVDTKDE EAEYQCDIEG CSMGFSSKQE LVLHKRNICS VKGCGKKFFS HKYLVQHRRV 1320 HMDDRPLKCP WKGCKMTFKW AWARTEHIRV HTGARPYVCR EPGCGQTFRF VSDFSRHKRK 1380 TGHSVKKGKG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 2e-79 | 28 | 460 | 8 | 413 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-79 | 28 | 460 | 8 | 413 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
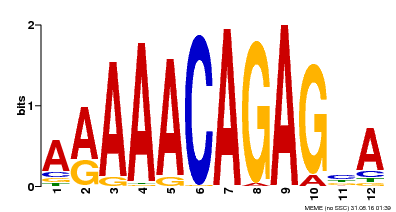
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM466832 | 5e-48 | AM466832.1 Vitis vinifera, whole genome shotgun sequence, contig VV78X011128.17, clone ENTAV 115. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010250905.1 | 0.0 | PREDICTED: lysine-specific demethylase JMJ705-like | ||||
| TrEMBL | A0A1U7ZHC1 | 0.0 | A0A1U7ZHC1_NELNU; lysine-specific demethylase JMJ705-like | ||||
| STRING | XP_010250905.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



