 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_004698-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 374aa MW: 43238.1 Da PI: 8.3601 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 20.9 | 9.7e-07 | 66 | 87 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C+ Cg sF+ + +Lk+H+++H
NNU_004698-RA 66 TCEECGASFRKPAYLKQHMQSH 87
6*******************99 PP
| |||||||
| 2 | zf-C2H2 | 19.2 | 3.2e-06 | 93 | 117 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp dC+ s++rk++L+rH+ H
NNU_004698-RA 93 FTCPvdDCSSSYRRKDHLTRHLLQH 117
89*******************9887 PP
| |||||||
| 3 | zf-C2H2 | 17.7 | 9.8e-06 | 122 | 147 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp +C+++Fs + n+krH + H
NNU_004698-RA 122 FTCPmeNCNRMFSLQGNMKRHVNEmH 147
89*******************99888 PP
| |||||||
| 4 | zf-C2H2 | 21.6 | 5.6e-07 | 162 | 184 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ Cgk+F+ s L++H+ +H
NNU_004698-RA 162 HACEECGKVFRFASKLRKHMDSH 184
67******************999 PP
| |||||||
| 5 | zf-C2H2 | 16.6 | 2.2e-05 | 251 | 275 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C +Fs+ksnL++Hi+ H
NNU_004698-RA 251 KCSfkGCLHTFSTKSNLNQHIKAvH 275
688889***************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 38 | 21 | 44 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 6.8E-8 | 64 | 86 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.33E-11 | 64 | 117 | No hit | No description |
| PROSITE profile | PS50157 | 13.464 | 65 | 92 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0039 | 65 | 87 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 67 | 87 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.9E-12 | 87 | 119 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.889 | 93 | 122 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0028 | 93 | 117 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 95 | 117 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-6 | 120 | 143 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 9.92E-8 | 120 | 184 | No hit | No description |
| PROSITE profile | PS50157 | 12.217 | 122 | 152 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0037 | 122 | 147 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 124 | 147 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-5 | 162 | 184 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.635 | 162 | 189 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0051 | 162 | 184 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 164 | 184 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.7 | 192 | 217 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 194 | 217 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 8.7 | 220 | 241 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 3.13E-7 | 231 | 292 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 5.5E-6 | 231 | 272 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.385 | 250 | 280 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0017 | 250 | 275 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 252 | 275 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.0E-5 | 274 | 304 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 1.1 | 281 | 307 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 8.767 | 281 | 307 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 283 | 307 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 374 aa Download sequence Send to blast |
MEGGDKEGAF TGPIFRDIRR YYCEFCGICR SKKSLITSHI LTYHKDEMEE KRDNNDQEKE 60 KRKSNTCEEC GASFRKPAYL KQHMQSHSIE RPFTCPVDDC SSSYRRKDHL TRHLLQHQGK 120 LFTCPMENCN RMFSLQGNMK RHVNEMHDDE EPSFSEGDGQ KHACEECGKV FRFASKLRKH 180 MDSHVKLDTV EAFCSEPGCL KCFTNVECLK AHIQSCHQYI TCEVCGVEQL KKNIKRHMRT 240 HEAGVTSESI KCSFKGCLHT FSTKSNLNQH IKAVHLELRP FACRFPGCGK KFPYKHVRDN 300 HEKTGTHVYV QGDFVESDEL FRTRPRGGRK RKCPAVETLL RKRVLPPSQM DSALSQGSDY 360 LAWLLSSTTH DEEQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2i13_A | 1e-16 | 63 | 247 | 20 | 190 | Aart |
| 2i13_B | 1e-16 | 63 | 247 | 20 | 190 | Aart |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 323 | 331 | RPRGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
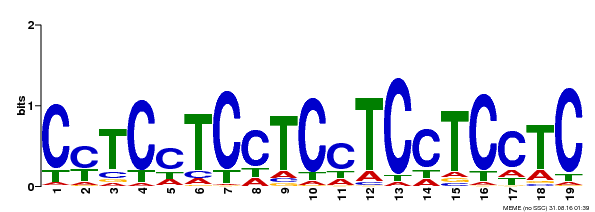
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010251838.1 | 0.0 | PREDICTED: transcription factor IIIA | ||||
| Refseq | XP_010251839.1 | 0.0 | PREDICTED: transcription factor IIIA | ||||
| Refseq | XP_010251840.1 | 0.0 | PREDICTED: transcription factor IIIA | ||||
| Refseq | XP_010251841.1 | 0.0 | PREDICTED: transcription factor IIIA | ||||
| Swissprot | Q84MZ4 | 1e-140 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A1U7ZTZ8 | 0.0 | A0A1U7ZTZ8_NELNU; transcription factor IIIA | ||||
| STRING | XP_010251838.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-142 | transcription factor IIIA | ||||



