 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_006931-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 285aa MW: 32200.6 Da PI: 9.1956 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 105.8 | 2.5e-33 | 21 | 75 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWtp+LH+rFv+av++LGG++kAtPk++l++m++kgLtl+h+kSHLQkYRl
NNU_006931-RA 21 KPRLRWTPDLHDRFVDAVTKLGGPDKATPKSVLRVMGLKGLTLYHLKSHLQKYRL 75
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.781 | 18 | 78 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-32 | 18 | 76 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-24 | 21 | 76 | IPR006447 | Myb domain, plants |
| SuperFamily | SSF46689 | 4.3E-16 | 21 | 77 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.2E-9 | 23 | 74 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 1.4E-24 | 125 | 171 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 285 aa Download sequence Send to blast |
MERTCGSYAL ETGMVLSRDP KPRLRWTPDL HDRFVDAVTK LGGPDKATPK SVLRVMGLKG 60 LTLYHLKSHL QKYRLGKQSK KGTNIEQAKS EELSVDSVGD SGYNHMHFSA TSSSTSKENS 120 ERGIPIAEAL RYQIEVQRRL QEQLEVQKKL QIRIEAQGKY LQAILEKAQK SLSTDTKCPR 180 SLEATRAQLT DFNLALSGLM ENMNQVREDE SKGEMIGKSL LNDNQENNHS SGFQLYRELE 240 EAKKKEIKIK VEESSLLFDL NVKGGYDFLG PKGSELDPNM NIQRI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 2e-19 | 21 | 77 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-19 | 21 | 77 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-19 | 21 | 77 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-19 | 21 | 77 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
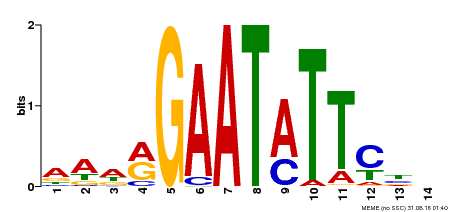
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010255718.1 | 0.0 | PREDICTED: myb family transcription factor PHL11 isoform X1 | ||||
| Refseq | XP_010255719.1 | 0.0 | PREDICTED: myb family transcription factor PHL11 isoform X1 | ||||
| Swissprot | C0SVS4 | 3e-79 | PHLB_ARATH; Myb family transcription factor PHL11 | ||||
| TrEMBL | A0A1U8A238 | 0.0 | A0A1U8A238_NELNU; myb family transcription factor PHL11 isoform X1 | ||||
| STRING | XP_010255718.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 3e-65 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



