 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_007725-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 588aa MW: 64376.5 Da PI: 6.4127 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 109.4 | 1.6e-34 | 239 | 297 | 1 | 60 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
ldDgynWrKYGqK+vkgse+prsYY+Ct+++Cpvkkkvers d++++ei+Y+g+Hnh+k
NNU_007725-RA 239 LDDGYNWRKYGQKQVKGSENPRSYYKCTFPDCPVKKKVERSL-DGQITEIVYKGNHNHPK 297
59****************************************.***************85 PP
| |||||||
| 2 | WRKY | 53.6 | 4.6e-17 | 423 | 461 | 21 | 59 |
-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 21 prsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+sYY+Ct+ gCpv+k+ver+++d ++v++tYeg+Hnh+
NNU_007725-RA 423 RKSYYKCTHVGCPVRKHVERASHDLRAVITTYEGKHNHD 461
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.4E-29 | 229 | 298 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.57 | 234 | 298 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.71E-26 | 234 | 298 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.7E-38 | 239 | 297 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 7.3E-26 | 240 | 296 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.5E-12 | 415 | 462 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.27E-14 | 420 | 463 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.9E-15 | 420 | 463 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.0E-12 | 422 | 461 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 21.597 | 424 | 463 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 588 aa Download sequence Send to blast |
MASSSGSLET SANSHPTPFS FSNQFMTSSF SDLLAGDDDK WLSDHFADRT GASVPKFKSI 60 PPPSLPISPP PVSPSSYFAI PPGLSPTDLL DSPVLLSASN LLQSPTTGTL PSQAFNWKSN 120 SVNYQHGIKK EEKNLPDFSF QPQTRPAMGS VSMFESSITT ISKDENSKGL EQEWNFQPTK 180 QADFSSGKPM VKSEYAPMQN FSHEISSIQA NNTQSNGGFQ SNHNQYSQPC QSFREQRRLD 240 DGYNWRKYGQ KQVKGSENPR SYYKCTFPDC PVKKKVERSL DGQITEIVYK GNHNHPKPQS 300 TRRSSSSSHR IQASAAPISE IPDPSFATRG VASQIMESGA TPENSSISMG EDDFEQSSQM 360 SRSAGDEFDE DEPNAKRWKT TTEGDQNEGI SAAGSRTVRE PRVIVQTTSD IDILDDDGEN 420 MGRKSYYKCT HVGCPVRKHV ERASHDLRAV ITTYEGKHNH DVPAARGSGS HLINRPTPDN 480 KNNNTAAAAN NVAMAAIRPT PTTNTFSPYS STNPLRSVRP PITAETQPPX TLEMLRSSHS 540 SPGNFGFSSG IGNSMPSYTN QSLHIDSSVF ATAKEEPGDD LFLESLLC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 8e-17 | 239 | 463 | 2 | 74 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor (By similarity). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (By similarity). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
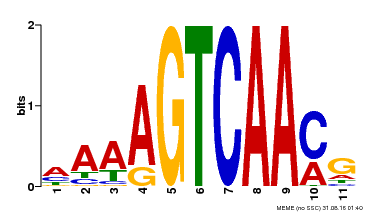
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:25110688). Slightly down-regulated by gibberellic acid (GA) (By similarity). Accumulates in response to jasmonic acid (MeJA) (PubMed:16919842). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000269|PubMed:16919842, ECO:0000269|PubMed:25110688}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010257005.1 | 0.0 | PREDICTED: WRKY transcription factor WRKY24-like isoform X2 | ||||
| Swissprot | Q6B6R4 | 1e-117 | WRK24_ORYSI; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A1U8A6I3 | 0.0 | A0A1U8A6I3_NELNU; WRKY transcription factor WRKY24-like isoform X2 | ||||
| STRING | XP_010257004.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 6e-82 | WRKY DNA-binding protein 33 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



