 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_009076-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 929aa MW: 105300 Da PI: 7.5812 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 165.3 | 1e-51 | 29 | 145 | 2 | 117 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
++e ++rwl+++ei+aiL n++ +++ ++ + p+sg++iL++rk++r+frkDG++wkkkkdgktv+E+he+LKvg e +++yYah+e+np+f r
NNU_009076-RA 29 MEEaRTRWLRPNEIHAILCNHTYFTVNVKPINLPQSGTIILFDRKVLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGDEERIHVYYAHGEDNPNFVR 124
45669******************************************************************************************* PP
CG-1 97 rcywlLeeelekivlvhylev 117
rcywlL++++e+ivlvhy+e+
NNU_009076-RA 125 RCYWLLDKKQEHIVLVHYRET 145
******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 76.637 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 5.1E-75 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.4E-45 | 31 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.9E-13 | 378 | 464 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 1.1E-16 | 560 | 675 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.28E-15 | 562 | 672 | No hit | No description |
| Pfam | PF12796 | 2.7E-6 | 562 | 642 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 9.79E-16 | 567 | 675 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 16.626 | 568 | 675 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.701 | 613 | 645 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 6.0E-6 | 613 | 642 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3500 | 652 | 681 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 58 | 757 | 779 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.272 | 761 | 787 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.108 | 777 | 806 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0014 | 799 | 821 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.823 | 800 | 824 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.6E-4 | 801 | 821 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 13 | 882 | 904 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.334 | 884 | 912 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 929 aa Download sequence Send to blast |
MERSVPGRLA GSEIHGFRTM EDLDVPSMME EARTRWLRPN EIHAILCNHT YFTVNVKPIN 60 LPQSGTIILF DRKVLRNFRK DGHNWKKKKD GKTVKEAHEH LKVGDEERIH VYYAHGEDNP 120 NFVRRCYWLL DKKQEHIVLV HYRETLEAQG SPVTPVNSNS SPENSGPFAS RVLSEENDSG 180 ANHGFYAGSG SPLVSESAEL DDHFSVLHEI NTLEWEDLLG AQDASNPSPP KRGEVAHLEQ 240 QNLYELRGSL HSQGSFLPTN SLPTTLSSFR HPTEQMAKSA SIDIRPPNSG YVQTAGVISN 300 NQWKDFEKTD ESLNASFGNS LLTQDSFGRW MNCIISDSPG SIDNVQLQSS ISTTHETTLS 360 EITDHHHHTS TQGQVFSITD VSPSWAFSTE ETKVIMVGFF HAEYSHIAES NLLCVIGDVC 420 VPVEMIQVGV FRCMASPNNT GFVDLYLSLD GRTPISQVLT FEYRSPLIDN QGASQEDKCK 480 WKEFQIQLRL ARLLFSTNNS LSILSSKVLP NALKEAKKFA LMTSAIEKDW AYLIKSIGNS 540 GIPFLQAKDI LFELTLKNKL QEWLLERVAE GSKTTIRDTR GQGVIHLCAI LGYTWAVYPY 600 SRSGLSLDFR DAYGWTALHW AAFYGREKMV AVLLSAGAKP NLVTDPTPEF PGGRTAADLA 660 SKNGYEGLSA YLAEKALIFQ FYEMKISGNA SGSLGTNTTT YTSPEALNED ELCLKDTLAA 720 YRTAADAAAH IQAAFRQHSL KLKEKAVQLA NPEMEARNII AAMKIQHAFR NYETRKKMTA 780 AARIQYRFRT WKIRKDFLNM RRQAIKIQAV FRGYQVRRQY RKILWSVGVL EKVILRWRLK 840 RKGFRGLSVE LEPTQEMPVD QNQESDVEDD FFRVSRKQAE ERVERSVVRV QAMFRSKQAQ 900 QEYRRMKLAY DQAALEYEDL LDPEVRNQK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
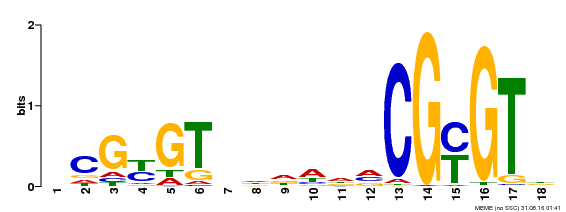
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010259339.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A1U8A3G4 | 0.0 | A0A1U8A3G4_NELNU; calmodulin-binding transcription activator 5 isoform X1 | ||||
| STRING | XP_010259339.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



