 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_011892-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 329aa MW: 36396.4 Da PI: 10.021 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 386.4 | 4.3e-118 | 1 | 329 | 1 | 301 |
GAGA_bind 1 mdddgsre..rnkgyye...paaslkenlglqlmssiaerdakirernlalsekkaavaerdmaflqrdkalaernkalverdnkllalllvensl 91
mdd ++re r+k +y+ ++ + +++ +q+m+++aerda+i+ern+al+ekkaa+aerdmaflqrd+a+aern+al+erdn+++al+++en+l
NNU_011892-RA 1 MDDGSQREngRHKDQYKtvhSQWMIPQHQMKQVMALMAERDAAIQERNVALAEKKAALAERDMAFLQRDAAIAERNNALMERDNAIAALEYRENAL 96
8999998889****99965433566788999****************************************************************9 PP
GAGA_bind 92 a....salpvgvqvlsgtksidslqqlsepqledsavelreeeklealpieeaaeeakekkkkkkrqr..akkpkekkak...kkkkksekskkkv 178
+ ++p+g++v +gtk++++l + l++ ++++r+++++ea+p+++a+ ea++ ++ k++++ +++p+++ ++ k kk +e+ +k v
NNU_011892-RA 97 SgsgvPTCPPGCGVPRGTKHVHHLTH-----LTEAPYHARDMHITEAYPLSSAPAEAHKPRRAKRTKKesKAVPLKRSQQpssKGKKGGEDLNKLV 187
9988889***************9888.....99*******************99999988777644430033444444433334444444444444 PP
GAGA_bind 179 kkesader.......................skaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrr 251
+ + + sk ++k +dl+ln+v++Dest+PvPvCsCtG+l+qCYkWGnGGWqSaCCttt+S yPLPv++++r
NNU_011892-RA 188 TVA----KlhdwrsgqdangsgedlkkqlavSKPDWKGQDLGLNQVNFDESTMPVPVCSCTGVLQQCYKWGNGGWQSACCTTTLSSYPLPVMPNKR 279
444....1445679********************************************************************************** PP
GAGA_bind 252 gaRiagrKmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
+aRi+grKmS++af+klL++Laa+G+dls+pvDLkdhWAkHGtn++vti+
NNU_011892-RA 280 HARIGGRKMSGSAFTKLLSRLAAQGHDLSTPVDLKDHWAKHGTNRYVTIK 329
*************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 2.0E-162 | 1 | 329 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 1.1E-102 | 1 | 329 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 329 aa Download sequence Send to blast |
MDDGSQRENG RHKDQYKTVH SQWMIPQHQM KQVMALMAER DAAIQERNVA LAEKKAALAE 60 RDMAFLQRDA AIAERNNALM ERDNAIAALE YRENALSGSG VPTCPPGCGV PRGTKHVHHL 120 THLTEAPYHA RDMHITEAYP LSSAPAEAHK PRRAKRTKKE SKAVPLKRSQ QPSSKGKKGG 180 EDLNKLVTVA KLHDWRSGQD ANGSGEDLKK QLAVSKPDWK GQDLGLNQVN FDESTMPVPV 240 CSCTGVLQQC YKWGNGGWQS ACCTTTLSSY PLPVMPNKRH ARIGGRKMSG SAFTKLLSRL 300 AAQGHDLSTP VDLKDHWAKH GTNRYVTIK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. {ECO:0000269|PubMed:14731261}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00540 | DAP | Transfer from AT5G42520 | Download |
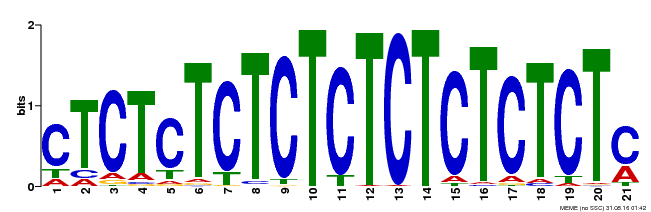
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010263912.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263913.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263915.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263916.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263917.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263918.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263919.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263920.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263921.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263922.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Refseq | XP_010263924.1 | 0.0 | PREDICTED: protein BASIC PENTACYSTEINE6 | ||||
| Swissprot | Q8L999 | 1e-133 | BPC6_ARATH; Protein BASIC PENTACYSTEINE6 | ||||
| TrEMBL | A0A1U8AMF6 | 0.0 | A0A1U8AMF6_NELNU; protein BASIC PENTACYSTEINE6 | ||||
| STRING | XP_010263912.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G42520.1 | 1e-120 | basic pentacysteine 6 | ||||



