 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_012716-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 365aa MW: 40798 Da PI: 9.0655 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 44.9 | 2.9e-14 | 61 | 109 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f +eeAa+a++ a+++++g
NNU_012716-RA 61 SQYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEEEAARAYDIAAQRFRG 109
78****9888.8*********3.....5*************************98 PP
| |||||||
| 2 | B3 | 101.2 | 6.1e-32 | 193 | 292 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE.. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefel 92
f+k+ tpsdv+k++rlv+pk++ae+h g++ + + l +ed g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F+++ +++l
NNU_012716-RA 193 FDKAVTPSDVGKLNRLVIPKQHAEKHfplqIGSTSKCVLLNFEDNGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDVVSFQRSTGPDKQL 288
89***************************945566889*************************************************876677777 PP
EEEE CS
B3 93 vvkv 96
++ +
NNU_012716-RA 289 YIDW 292
7766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 9.81E-18 | 61 | 117 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.11E-27 | 61 | 116 | No hit | No description |
| PROSITE profile | PS51032 | 20.521 | 62 | 117 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.6E-9 | 62 | 109 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.7E-21 | 62 | 117 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.2E-30 | 62 | 123 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.5E-5 | 63 | 74 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.5E-5 | 83 | 99 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 1.2E-40 | 186 | 300 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.7E-29 | 191 | 291 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.07E-27 | 192 | 281 | No hit | No description |
| Pfam | PF02362 | 1.2E-28 | 193 | 292 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.099 | 193 | 296 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.3E-24 | 193 | 296 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 365 aa Download sequence Send to blast |
MEGSCIDEST SDSISVTPLP VLALPGKKEP EKLCRVGSGA SSVVLDSESG VEAESRKLPS 60 SQYKGVVPQP NGRWGAQIYE KHQRVWLGTF NEEEEAARAY DIAAQRFRGR DAVTNFKPLS 120 ETDEDDIETI FLNSHSKAEI VDMLRKHTYH DELEQTKRNY GCNGDGSRRI RGGGGFGDST 180 GSDRVAKARE QLFDKAVTPS DVGKLNRLVI PKQHAEKHFP LQIGSTSKCV LLNFEDNGGK 240 VWRFRYSYWN SSQSYVLTKG WSRFVKEKNL KAGDVVSFQR STGPDKQLYI DWKPRAASTY 300 MALPQLVHPV NPVQPVEVVR LFGVNIFKIP VSSVVDTWNI NGKRLRDVDF SSLESCKKQR 360 VVGAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 1e-55 | 190 | 298 | 11 | 120 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences. Functionally redundant with TEM2. {ECO:0000269|PubMed:18718758}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00024 | PBM | Transfer from AT1G25560 | Download |
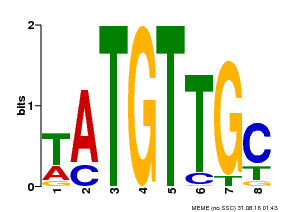
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Expressed with a circadian rhythm showing a peak at dawn. {ECO:0000269|PubMed:18718758}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010265251.1 | 0.0 | PREDICTED: AP2/ERF and B3 domain-containing transcription factor RAV1-like | ||||
| Swissprot | Q9C6M5 | 1e-148 | RAVL1_ARATH; AP2/ERF and B3 domain-containing transcription repressor TEM1 | ||||
| TrEMBL | A0A1U8AE70 | 0.0 | A0A1U8AE70_NELNU; AP2/ERF and B3 domain-containing transcription factor RAV1-like | ||||
| STRING | XP_010265251.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13260.1 | 1e-145 | related to ABI3/VP1 1 | ||||



