 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_012928-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 497aa MW: 55633.9 Da PI: 4.5641 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 173.8 | 1.8e-53 | 96 | 207 | 1 | 120 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWk 96
+el++rmwkd+++l+r+ker+k ++ ++ k +++++qarrkkmsraQDgiLkYMlk mevcna+GfvYgi+pekgkpv+g+sd++raWWk
NNU_012928-RA 96 DELERRMWKDRVKLNRIKERQKLAA--QQ---EKPKPKQTSDQARRKKMSRAQDGILKYMLKLMEVCNARGFVYGIVPEKGKPVSGSSDNIRAWWK 186
79*****************999644..33...467999********************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 97 ekvefdrngpaaiskyqaknlils 120
ekv+fd+ngpaai+k+qa ++s
NNU_012928-RA 187 EKVKFDKNGPAAIAKKQA---VAS 207
***************965...444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 3.4E-53 | 96 | 207 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 497 aa Download sequence Send to blast |
MVNFNTAWAA ALQTCGQKLT KMLCEASGIR DVSAASWKAE EEEGGEGVIT KLVCRLKEMA 60 DLEEIGAEIC SDLEVDDIRC DNLAEKDVSD EEIEADELER RMWKDRVKLN RIKERQKLAA 120 QQEKPKPKQT SDQARRKKMS RAQDGILKYM LKLMEVCNAR GFVYGIVPEK GKPVSGSSDN 180 IRAWWKEKVK FDKNGPAAIA KKQAVASSDS DYDVDGVDDG VGSVSSNPKV DRRIQPVNAG 240 PCGNPCDNPR HPSNDKEQSE EQPRRKRPRG RSKPVDQGTP SSQNEHLHGE ERSVVPDMNQ 300 VDLQLVEFQM QTTQLENDPV PSLRPLEEDQ DIQLQLPESA LNTFSDLPST NETTSESMLV 360 GDRPLLYSIV QNAELPPRPS YFFYDPSVGN RLHQDKEQLQ ISMADPQMMP EETGYHAPLL 420 HGNRNQIAGE EIHRYGKDSF HSEQDKPVES HYGSPLDGLS LDYGGFNSPF FGFDGTSSLD 480 ADLDFSLDDD LIQYFGA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
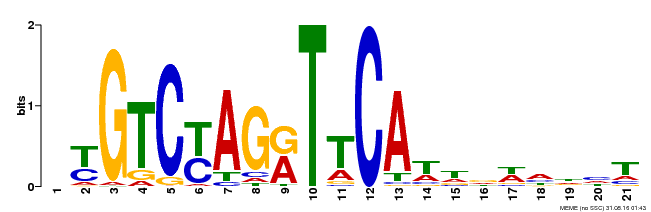
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019054338.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 5e-64 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A1U8Q6A8 | 0.0 | A0A1U8Q6A8_NELNU; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| STRING | XP_010265828.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-66 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



