 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_014544-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 702aa MW: 76205.9 Da PI: 5.7413 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 91.3 | 8.6e-29 | 213 | 266 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv av+qL G++kA+Pk+ilelm+v+gLt+e+v+SHLQkYRl
NNU_014544-RA 213 KPRVVWSVELHQQFVAAVNQL-GIDKAVPKKILELMNVPGLTRENVASHLQKYRL 266
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 77.2 | 5.7e-26 | 35 | 143 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTE CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGa 94
vl+vdD+p+ +++l ++l++ y ev+ +d +e al ll+e++ +D+++ D+ mp+mdG++ll++i e +lp+i+++a + ++ + + + Ga
NNU_014544-RA 35 VLVVDDDPTCLRILDKMLRNCLY-EVTICDRAEVALSLLRESKsgFDIVISDVHMPDMDGFKLLEHIGLEM-DLPVIMMSADDGKDVVMKGVTHGA 128
89*********************.***************999999**********************6654.8*********************** PP
SEEEESS--HHHHHH CS
Response_reg 95 kdflsKpfdpeelvk 109
d+l Kp+ +e+l +
NNU_014544-RA 129 CDYLIKPVRIEALKN 143
***********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 2.0E-172 | 1 | 692 | IPR017053 | Response regulator B-type, plant |
| Gene3D | G3DSA:3.40.50.2300 | 2.8E-41 | 32 | 175 | No hit | No description |
| SuperFamily | SSF52172 | 7.83E-35 | 32 | 157 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 5.2E-29 | 33 | 145 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.035 | 34 | 149 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 9.0E-23 | 35 | 143 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 4.36E-27 | 36 | 148 | No hit | No description |
| SuperFamily | SSF46689 | 3.76E-20 | 210 | 270 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.048 | 210 | 269 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-30 | 212 | 271 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.0E-24 | 213 | 266 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.0E-7 | 215 | 265 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 702 aa Download sequence Send to blast |
MNMDSGGGSV SLANSILTKK AADAISDQFP AGLRVLVVDD DPTCLRILDK MLRNCLYEVT 60 ICDRAEVALS LLRESKSGFD IVISDVHMPD MDGFKLLEHI GLEMDLPVIM MSADDGKDVV 120 MKGVTHGACD YLIKPVRIEA LKNIWQHVVR KRRNDLKELE QSGSMEDSDR HRKPSEDGDY 180 ASSVNDGNWR NSKRRKDEEE EGEERDDTST LKKPRVVWSV ELHQQFVAAV NQLGIDKAVP 240 KKILELMNVP GLTRENVASH LQKYRLYLRR LSGVSQHQNG LNTPFMGPQE ASFGPMSTLD 300 GLDLQALAVS GQLAPQSLAT LQAGGLGRST ANASISMPLV DQRNIFTSDA SKLRVGAGQQ 360 SVNSSSKQVN LIHGLPTNME PKQLSHLHQS VQSFGTMGLQ VSEGVSSFLN IPATSLRTSS 420 SHVDSVLGSQ NSSLMIQMAQ PQSRGHLLND ITGSHSSGLQ SSMVQQIFSN EIAGQVLGRN 480 GVILNGKGAA YNLVSQASPM ADFPLNHIAE LPGNGLPIGN TLTSSSTLTS TGMFQGGNVD 540 TKGPRGFAPV CDIFSEICQS KPQDWELQNV GLTFEASQPI TSVQGNVDLT QAVLGNQGFA 600 SSQKNLLNRN ASVVGKAVVP QRQENEHGKI EASVQRPINP LLNNSFRVKP ETISDFSCDN 660 LLIPQRYGQD DLMSALFKQQ EGTGSLESEC NFDGYTMDNI PV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-22 | 212 | 272 | 4 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins. Involved in the expression of nuclear genes for components of mitochondrial complex I. Promotes cytokinin-mediated leaf longevity. Involved in the ethylene signaling pathway in an ETR1-dependent manner and in the cytokinin signaling pathway. {ECO:0000269|PubMed:11370868, ECO:0000269|PubMed:11574878, ECO:0000269|PubMed:15282545, ECO:0000269|PubMed:16407152}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
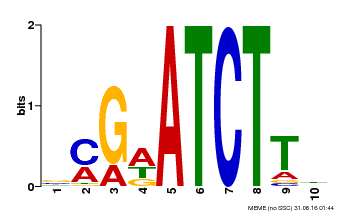
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010268091.1 | 0.0 | PREDICTED: two-component response regulator ARR2-like isoform X2 | ||||
| Swissprot | Q9ZWJ9 | 0.0 | ARR2_ARATH; Two-component response regulator ARR2 | ||||
| TrEMBL | A0A1U8APJ3 | 0.0 | A0A1U8APJ3_NELNU; two-component response regulator ARR2-like isoform X2 | ||||
| STRING | XP_010268090.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-176 | response regulator 2 | ||||



