 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_014660-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 706aa MW: 78101.7 Da PI: 6.2561 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.5 | 7.2e-21 | 22 | 77 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t+ q+++Le++F+++++p++++r +L++ lgL+ rq+k+WFqNrR+++k
NNU_014660-RA 22 KKRYHRHTARQIQALEAMFKECPHPDEKQRMQLSRDLGLEPRQIKFWFQNRRTQMK 77
688899***********************************************998 PP
| |||||||
| 2 | START | 174.7 | 5.6e-55 | 219 | 442 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE.......EXCCTTEEEEEEESSS...SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEE CS
START 2 laeeaaqelvkkalaeepgWvkss.......esengdevlqkfeeskv..dsgealrasgvvdmvlallveellddkeqWdetla....kaetlev 84
+a a++el+++ + +ep+W+ks + en++ + ++ + k+ ++ea+r+sgvv+m+ lv +++++ +W e ++ ka t+ev
NNU_014660-RA 219 IAANALDELIRLLQTDEPLWMKSTtdgrdvlDLENYERTFPRPNRMKNpnVQIEASRDSGVVIMNGLALVDMFMNSN-KWVELFPtivsKARTVEV 313
788999******************999977766666666666666555999**************************.******99999******* PP
ECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE-- CS
START 85 issg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvdlk 173
+ssg g lql++aelq+lsplv+ R+ +f+Ry++q ++g w+ivdvS ds ++++ s+ +++lpSg+li++++ng+sk+ wveh++l+
NNU_014660-RA 314 LSSGmignrsGSLQLIYAELQVLSPLVStRELYFLRYCEQIEQGLWAIVDVSFDSSRENQ-FLSSSHCRRLPSGCLIQDMPNGYSKISWVEHMELE 408
***********************************************************9.68888889*************************** PP
SSXX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 174 grlp.hwllrslvksglaegaktwvatlqrqcek 206
+++ h l+r lv sg+a+ga +w+atlqr ce+
NNU_014660-RA 409 EKTStHRLYRDLVYSGMAFGAERWLATLQRMCER 442
**999***************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.6E-20 | 15 | 82 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-23 | 17 | 83 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.734 | 19 | 79 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.6E-19 | 20 | 83 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.96E-18 | 21 | 80 | No hit | No description |
| Pfam | PF00046 | 2.2E-18 | 22 | 77 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 54 | 77 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 47.882 | 209 | 445 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.13E-34 | 210 | 443 | No hit | No description |
| CDD | cd08875 | 4.17E-121 | 213 | 441 | No hit | No description |
| SMART | SM00234 | 1.5E-47 | 218 | 442 | IPR002913 | START domain |
| Pfam | PF01852 | 2.4E-47 | 219 | 442 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 4.1E-5 | 271 | 399 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 1.15E-21 | 464 | 698 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 706 aa Download sequence Send to blast |
MESDTRGSGS GEDLDASDPQ RKKRYHRHTA RQIQALEAMF KECPHPDEKQ RMQLSRDLGL 60 EPRQIKFWFQ NRRTQMKAQH ERADNCALRA ENDKIRCENI AIREALKNVI CPSCGGPPVG 120 DDMYFDEQKL RMENARLREE LDRVSSIAAK YIGRPISQLP TIQPIPVSSL DLSVGSFGGQ 180 GMVGPSLDLD LLPGSSSVPQ FPFQTVVISE MDKPLMAEIA ANALDELIRL LQTDEPLWMK 240 STTDGRDVLD LENYERTFPR PNRMKNPNVQ IEASRDSGVV IMNGLALVDM FMNSNKWVEL 300 FPTIVSKART VEVLSSGMIG NRSGSLQLIY AELQVLSPLV STRELYFLRY CEQIEQGLWA 360 IVDVSFDSSR ENQFLSSSHC RRLPSGCLIQ DMPNGYSKIS WVEHMELEEK TSTHRLYRDL 420 VYSGMAFGAE RWLATLQRMC ERFACQMVTS TSARDLGGVI PSAEGRRSMM KLAQRMINSF 480 CASISASNGH RWTTLSGLND IGVRVTVHKS TEAGQPNGVV LSATTSIWLP VSPQNVFNFF 540 RDERTRAQWD VLSNGNSVQE VAHIANGSHP GNCISVLRAF NASQNNMLIL QESCTDSSGS 600 LVVYSPVDLT AINIAMSGED PSHISLLPSG FTISPDGQLD QVMGASTSSL QGSMSKSSGS 660 LITVAFQILV SSLPSAKLNL ESVTTVNNLI STTVQQIKAA LNCPSS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which acts as positive regulator of drought stress tolerance. Can transactivate CIPK3, NCED3 and ERECTA (PubMed:18451323). Transactivates several cell-wall-loosening protein genes by directly binding to HD motifs in their promoters. These target genes play important roles in coordinating cell-wall extensibility with root development and growth (PubMed:24821957). Transactivates CYP74A/AOS, AOC3, OPR3 and 4CLL5/OPCL1 genes by directly binding to HD motifs in their promoters. These target genes are involved in jasmonate (JA) biosynthesis, and JA signaling affects root architecture by activating auxin signaling, which promotes lateral root formation (PubMed:25752924). Acts as negative regulator of trichome branching (PubMed:16778018, PubMed:24824485). Required for the establishment of giant cell identity on the abaxial side of sepals (PubMed:23095885). May regulate cell differentiation and proliferation during root and shoot meristem development (PubMed:25564655). {ECO:0000269|PubMed:16778018, ECO:0000269|PubMed:18451323, ECO:0000269|PubMed:23095885, ECO:0000269|PubMed:24821957, ECO:0000269|PubMed:24824485, ECO:0000269|PubMed:25564655, ECO:0000269|PubMed:25752924}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
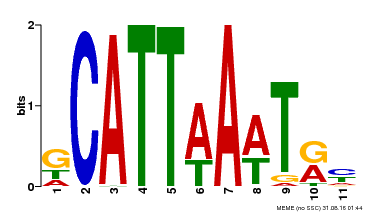
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010268321.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG11-like isoform X1 | ||||
| Swissprot | Q9FX31 | 0.0 | HDG11_ARATH; Homeobox-leucine zipper protein HDG11 | ||||
| TrEMBL | A0A1U8AQ32 | 0.0 | A0A1U8AQ32_NELNU; homeobox-leucine zipper protein HDG11-like isoform X1 | ||||
| STRING | XP_010268321.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



