 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_014892-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 273aa MW: 30026.2 Da PI: 7.8455 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 62.3 | 7.4e-20 | 37 | 98 | 2 | 57 |
T--SS--HHHHHHHHHHHHH..SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 2 rkRttftkeqleeLeelFek..nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
++R+t+t++q+++L++l+++ r psae++++++++l +++ ++V++WFqN++a+e++
NNU_014892-RA 37 STRWTPTTDQIRILKDLYYNngIRSPSAEQIQRISARLrqygKIEGKNVFYWFQNHKARERQ 98
58*****************8567*************************************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 113.7 | 9e-37 | 36 | 100 | 2 | 65 |
Wus_type_Homeobox 2 artRWtPtpeQikiLeelyk.sGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
++tRWtPt++Qi+iL++ly+ +G+r+P++e+iqri+a+L++yGkie+kNVfyWFQN+kaRerqk+
NNU_014892-RA 36 SSTRWTPTTDQIRILKDLYYnNGIRSPSAEQIQRISARLRQYGKIEGKNVFYWFQNHKARERQKK 100
789***************9779*****************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.07E-11 | 29 | 105 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 10.527 | 33 | 99 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.3E-5 | 35 | 103 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.9E-17 | 38 | 98 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-7 | 38 | 98 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 2.32E-7 | 38 | 100 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0080166 | Biological Process | stomium development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 273 aa Download sequence Send to blast |
MEPQQQPAQQ MQQQQQPNED SSSSNTSKSS FLCRQSSTRW TPTTDQIRIL KDLYYNNGIR 60 SPSAEQIQRI SARLRQYGKI EGKNVFYWFQ NHKARERQKK RFTVDMAMQR SVATGGWKTG 120 DPIHNKYTNI SPGVSSPSSP GVLAAGQMGG TGYGPVLMEK SFRDCSISAG GSGIGGGGMC 180 NNLGWVGIDP YSSPYALFDK RKSIEYLEDQ TPQIETLPLF PTHGENIGGF CTGKADSGGY 240 FTDWYGPSDE KNDTPTSLEL SLNSCTTTSP DSP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that plays a central role during developmental processes such as early embryogenesis and flowering, probably by regulating expression of specific genes. Required to specify stem cell identity in meristems, such as shoot apical meristem (SAM). Acts in parallel with HAM. {ECO:0000269|PubMed:12208843}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00265 | DAP | Transfer from AT2G17950 | Download |
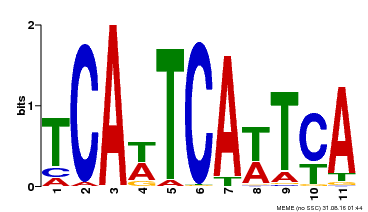
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM447418 | 9e-71 | AM447418.2 Vitis vinifera contig VV78X032979.11, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010268704.1 | 0.0 | PREDICTED: protein WUSCHEL | ||||
| Swissprot | Q8LL11 | 6e-95 | WUS_PETHY; Protein WUSCHEL | ||||
| TrEMBL | A0A1U8AMM3 | 0.0 | A0A1U8AMM3_NELNU; protein WUSCHEL | ||||
| STRING | XP_010268704.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G17950.1 | 1e-44 | WOX family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



