 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_015475-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 268aa MW: 29969.7 Da PI: 7.5035 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 63.9 | 3.3e-20 | 118 | 166 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+G+r+++ +g+W+AeIrdp++ r++lg++ taeeAa+a++ a+k+++g
NNU_015475-RA 118 YRGIRQRP-WGKWAAEIRDPRK---GVRVWLGTYNTAEEAARAYDVAAKRIRG 166
9*******.**********954...4************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 1.4E-31 | 117 | 175 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 5.82E-34 | 117 | 176 | No hit | No description |
| SMART | SM00380 | 1.2E-38 | 117 | 180 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.57E-22 | 117 | 174 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 24.171 | 117 | 174 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.1E-14 | 118 | 166 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.4E-11 | 118 | 129 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.4E-11 | 140 | 156 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008219 | Biological Process | cell death | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051707 | Biological Process | response to other organism | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 268 aa Download sequence Send to blast |
MDGCKCNGGI INEKACVVES LRIHSIFVSL SEPNMCGGAI ISDFIAVKRS RNLMSQDLWS 60 EFDTISDLLG FDFTDKDSSS SSTHQPSKGT RDKTKKRSAV GADQSKGKKK TAARKNIYRG 120 IRQRPWGKWA AEIRDPRKGV RVWLGTYNTA EEAARAYDVA AKRIRGDKAK LNFAEQTPPP 180 PKRRCLVPKS TQMSFEASGS PTTENEICEH NHHQPKDDME LREQISNLES FLELDPEPAE 240 SVGSVESEAF DLWLLDDVPF TPHGHLVY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 1e-21 | 118 | 173 | 3 | 59 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250, ECO:0000269|PubMed:9159183}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00109 | PBM | Transfer from AT3G16770 | Download |
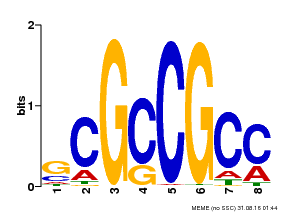
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By 1-aminocyclopropane-1-carboxylic acid (ACC, ethylene precursor), methyl jasmonate (MeJA), and Botrytis cinerea. Also induced by cadmium. {ECO:0000269|PubMed:18836139, ECO:0000269|PubMed:9159183}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010269611.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-3-like | ||||
| Swissprot | P42736 | 2e-56 | RAP23_ARATH; Ethylene-responsive transcription factor RAP2-3 | ||||
| TrEMBL | A0A1U8AT69 | 0.0 | A0A1U8AT69_NELNU; ethylene-responsive transcription factor RAP2-3-like | ||||
| STRING | XP_010269611.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G16770.1 | 3e-49 | ethylene-responsive element binding protein | ||||



