 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_019690-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1023aa MW: 113423 Da PI: 6.9192 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 130.7 | 5.1e-41 | 160 | 237 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C adls+ak+yhrrhkvCe+hska+++lv++++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
NNU_019690-RA 160 VCQVKDCGADLSNAKDYHRRHKVCEMHSKASKALVDNVMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTHP 237
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.8E-33 | 155 | 222 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.223 | 158 | 235 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.58E-38 | 159 | 240 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.7E-30 | 161 | 234 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 4.97E-9 | 789 | 906 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 5.4E-7 | 791 | 907 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.84E-8 | 795 | 905 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1023 aa Download sequence Send to blast |
MEARIGGEFH HFYGQGASGI VLEEKDWMGV GRRSLEWDLN DWKWDGDLFI SSPLNSVPSD 60 CRGRHLFPGS SGIPTAGGSS NSSSSCSDEI NPGSEKGKRE LEKRRRVIVV EDEELNDEAG 120 SLTLKLGGHG YPITEADISN WDGKNGKKTK LLGTTSNRAV CQVKDCGADL SNAKDYHRRH 180 KVCEMHSKAS KALVDNVMQR FCQQCSRFHV LQEFDEGKRS CRRRLAGHNK RRRKTHPETV 240 VNGSSLNDEQ ATNSSDQTKD QDLLSHLLRN LASFAGAVDG RNISGLLQDS QDPLKVGTSV 300 GKSSEKVAPL LTNGADTTRL VGSTSKINCN GAQGPQIGSS DHCFGASTAV MPQKVMVTED 360 ARVGVLQVVS SQKSTTLFPM KHGNPSKGTQ SMARRTKLNN IDLNNIYNDS QDCIEDAEGS 420 QAPALDFPTW MQQDSHQSSP PQASRNSDSA SAQSPSSSSG DTQSRTDRIV FKLFGKDPSD 480 FPLVLRAQIV DWLSHSPTDM ESYIRPGCII LTVYLRLQDS TWDEICGDLS SSLSRLLDAS 540 DGSFWRTGWV YARVQHRIAF VYNGQIVLDT PLPLKTHNHC RISSIAPIAV TVSEKARFIV 600 KGFNLSRPTT RLLCALEGNY LVQEATRDLV VSNEIFKEQD EFQCLRFDSS IPDVIGRGFI 660 EVEDHGLSSS FFPFIVAEQD VCSEIRMLES VIEEAKSEND AQRRTEKIEA KNQALDFIHE 720 MGWLLHRTHM RSRLGHMDPN LDAFSFKRFR LIMEFSMDHG WCAVVKKLLD IVFKGNVDVE 780 EHPSVELALS EMGLLHRAVR RNCKPLVELL LRYIPDNSDG AESKYNQQGV RVFDGFLFRP 840 DVVGPAGLTP LHVAASRDGC ENVLDALTDD PGMVGVDAWK NARDNTGFTP EDYARLRGHY 900 SYIHLVHKKA KKPEAGHVVL DIPGVLPDCN NNQKQMDGLP VGKGTSFQID KTKLAVSRYC 960 KACDQRLASY GTTSRSLVYR PAMLSMVAIA AVCVCVALLF KSSPEVLCVF PPFRWELLDY 1020 GPM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-30 | 160 | 234 | 10 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 102 | 106 | KRRRV |
| 2 | 229 | 235 | KRRRKTH |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
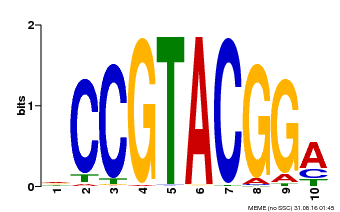
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010276543.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A1U8B6F9 | 0.0 | A0A1U8B6F9_NELNU; squamosa promoter-binding-like protein 1 | ||||
| STRING | XP_010276543.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



