 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_021919-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 752aa MW: 80543.3 Da PI: 7.0972 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.4e-16 | 478 | 524 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
NNU_021919-RA 478 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 524
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 2.5E-20 | 471 | 532 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.22E-20 | 472 | 535 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 474 | 523 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.81E-17 | 477 | 528 | No hit | No description |
| Pfam | PF00010 | 1.1E-13 | 478 | 524 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 480 | 529 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 752 aa Download sequence Send to blast |
MPLSEFHQMA KGKLESSQSK MNNCSTDQAF VPDHDFVELV WENGQILMQG QSSKTRRSPS 60 SNNLPSYISK VQDKDGGDAT NLKIGRSRTE EHVLNDFTST VPSGNMGLAS EEEMVPWLNY 120 SLDDSLQHDY CSEFFSELTG VNLNALSTQN NSFPIDKNSS CGQISRDLNI ASVYFDKNPQ 180 GNASKGVGGN PEPIRPSSNQ LRSSSLQQCQ ISLPSLRPRV SDLINSNTNN ADKASCGNSS 240 DTPTSNGGLP NTKMQRQDPG PTRPPPQNNI SSGLMNFSHF SRPAAFVKAN LHNLGAVASP 300 GLSSIDNLRS NGKASAAGSS NPVESTLIDS TSGSRNATSF HNQRASVPAE GEPKPSKPPK 360 EQFSAENSEV VFREEAHRRK RSPDRVIDHT SSFAASTAVG KPDGERAVEP AVASSSVCSV 420 NSGGGASDDP KNTLKRKSRD GDESEYQSED LEEESVGVKK LAPSRGGTSA KRSRAAEVHN 480 LSERRRRDRI NEKMRALQEL IPNCNKVDKA SMLDEAIEYL KTLQLQVQIM SMGSGLCMPP 540 MMLPPGMQHL HMPHLAHFSP MGVGMGMGMG MGLGFGMGMV DMSSGSSGRP LIQVPSMHGR 600 QFPCPPISGP ASLPGMAGAN LQMFGLPSQG LPMSIPRAPI IPLSGGSSTK SISLPDTSGT 660 VTPVKVPDSA PPSSSKEPEP NINPQMMHKT TADCSPDHIS TQILEGICIC TFTRDTSAVK 720 SCLNLVCPTC KCYGAQGMDA CHGYGIWYTE KV |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 482 | 487 | ERRRRD |
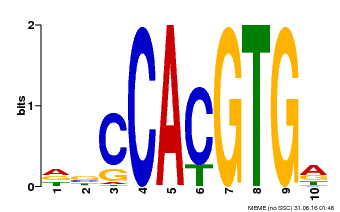
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010240811.1 | 0.0 | PREDICTED: transcription factor PIF3 isoform X2 | ||||
| TrEMBL | A0A1U7YT23 | 0.0 | A0A1U7YT23_NELNU; transcription factor PIF3 isoform X2 | ||||
| STRING | XP_010240809.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 2e-33 | phytochrome interacting factor 3 | ||||



