 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_024147-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 474aa MW: 50563.8 Da PI: 6.1973 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 80.4 | 2.1e-25 | 343 | 406 | 1 | 64 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHC CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkseve 64
e+elkr+rrkq+NRe+ArrsR+RK+ae+eeL+ kv++L++eN++L+ ele+l +e+akl+se++
NNU_024147-RA 343 ERELKRQRRKQSNRESARRSRLRKQAECEELQSKVQTLTSENHTLRTELERLAEECAKLTSENK 406
89***********************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 5.3E-34 | 66 | 160 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 2.9E-10 | 192 | 308 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 7.9E-19 | 338 | 402 | No hit | No description |
| SMART | SM00338 | 5.7E-24 | 343 | 407 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 3.5E-22 | 343 | 406 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.794 | 345 | 408 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.13E-12 | 346 | 402 | No hit | No description |
| CDD | cd14702 | 4.88E-24 | 348 | 399 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 350 | 365 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010310 | Biological Process | regulation of hydrogen peroxide metabolic process | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0090342 | Biological Process | regulation of cell aging | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 474 aa Download sequence Send to blast |
MERWNEAGEM GLVKTASGLW KMLTNPCRAS YLGNSTLVLV ILVFRVVDLI PWGIPDINVY 60 ILRSEMGSGE DSTPAKPSKP TTSAQETPTT PSYPDWSTPL QAYYGAGATA PPPFFTSSVA 120 SSPAPHPYLW GSQHLMPPYG TPLPYPALYP HGGLYAHPSM AAAQGAAVTT TEAEGKASDG 180 KDRSSAKKSK RSSGKESRKS ASGSANDGAS QSDESGNEGS SSASDENTNQ QDGKKRSFDQ 240 MLADGASAQN NSSAQYSGAT VETTFGTRGQ PLAKLPVSVP GRPVVNMPAA NLNIGMDLWN 300 SSPAGTVPLK TRPNASGVSA SVAQAPVVGR EGVMLDHLSI QDERELKRQR RKQSNRESAR 360 RSRLRKQAEC EELQSKVQTL TSENHTLRTE LERLAEECAK LTSENKSLME ELTQLYGPEA 420 IADLEDNNAS TVLQSTNGDV NGHQQDTSRE NDSACSQKNG TILNSNGKLD SGSS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 359 | 365 | RRSRLRK |
| 2 | 359 | 366 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the G-box motif (5'-CCACGTGG-3') of the rbcS-1A gene promoter (PubMed:1373374). G-box and G-box-like motifs are cis-acting elements defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control. Binds to the G-box motif 5'-CACGTG-3' of LHCB2.4 (At3g27690) promoter. May act as transcriptional activator in light-regulated expression of LHCB2.4. Probably binds DNA as monomer. DNA-binding activity is redox-dependent (PubMed:22718771). {ECO:0000269|PubMed:1373374, ECO:0000269|PubMed:22718771}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00039 | PBM | Transfer from AT4G36730 | Download |
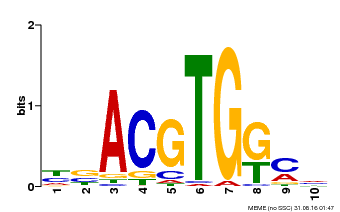
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010244323.1 | 0.0 | PREDICTED: G-box-binding factor 1-like isoform X2 | ||||
| Swissprot | P42774 | 5e-98 | GBF1_ARATH; G-box-binding factor 1 | ||||
| TrEMBL | A0A1U7Z163 | 0.0 | A0A1U7Z163_NELNU; G-box-binding factor 1-like isoform X2 | ||||
| STRING | XP_010244321.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36730.2 | 2e-75 | G-box binding factor 1 | ||||



