 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | NNU_024688-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Proteales; Nelumbonaceae; Nelumbo
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 356aa MW: 40085.5 Da PI: 8.5559 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.1 | 3.5e-32 | 179 | 232 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
NNU_024688-RA 179 PRMRWTTTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 232
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.74E-15 | 176 | 232 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-28 | 177 | 232 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.2E-23 | 179 | 232 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.3E-7 | 180 | 231 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 356 aa Download sequence Send to blast |
MELFPAQPDL SLQISPPNSK PTSGWKRTDE ELDLGFWRRA LDSNNTITSM AKSDTGFELS 60 LSNPRTSETN SNHLHHLLHH GHSNLIHANQ HHHHHHQNQQ NQYLHQQQAL QQELGFLKPI 120 RGIPVYHNLP SFPFVPQHTL DSSASTTTTT TSTSFPSHQS LMRSRFLSRF PAKRSMRAPR 180 MRWTTTLHAR FVHAVELLGG HERATPKSVL ELMDVKDLTL AHVKSHLQMY RTVKTTDRPA 240 ASGQPDGFDN GSAGEVSDDI MHESRGSELT VQQGRQSMHQ DQDYCGLWSN NSSSREAWLH 300 GKPRDSVGHT PPFEKEMDPK SLNYERISDL NSSSLTAGTG TSPKKPNLEF TLGRAV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 6e-16 | 180 | 234 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-16 | 180 | 234 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-16 | 180 | 234 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-16 | 180 | 234 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 6e-16 | 180 | 234 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that regulates lateral organ polarity. Promotes abaxial cell fate during lateral organd formation. Functions with KAN1 in the specification of polarity of the ovule outer integument. {ECO:0000269|PubMed:11525739, ECO:0000269|PubMed:15286295, ECO:0000269|PubMed:16623911, ECO:0000269|PubMed:17307928}. | |||||
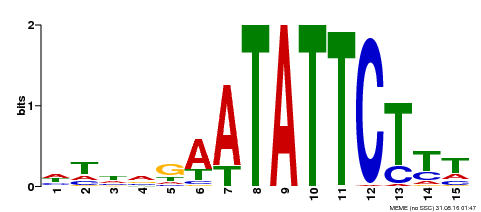
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010245225.1 | 0.0 | PREDICTED: probable transcription factor KAN2 isoform X1 | ||||
| Swissprot | Q9C616 | 4e-85 | KAN2_ARATH; Probable transcription factor KAN2 | ||||
| TrEMBL | A0A1U7ZBK8 | 0.0 | A0A1U7ZBK8_NELNU; probable transcription factor KAN2 isoform X1 | ||||
| STRING | XP_010245225.1 | 0.0 | (Nelumbo nucifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G16560.1 | 1e-50 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



