 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_11185_f_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 841aa MW: 94152.8 Da PI: 6.7975 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 71.6 | 1e-22 | 150 | 251 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrse 89
f+k+lt sd++++g +++ +++a+e+ + + + +++l+ +d++ ++W++++i+r++++r++l++GW+ Fv++++L +gD ++F + ++
Neem_11185_f_1 150 FCKTLTASDTSTHGGFSVLRRHADEClppldmSRQ-PPTQELVAKDLHANEWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL--RGEN 241
99*****************************7333.34459************************************************..4499 PP
E..EEEEE-S CS
B3 90 felvvkvfrk 99
+el+v+v+r+
Neem_11185_f_1 242 GELRVGVRRA 251
99*****996 PP
| |||||||
| 2 | Auxin_resp | 117.9 | 8.6e-39 | 276 | 358 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+st++ F+v+Y+Pr+s++eF+v+++++++++k+++s+GmRfkm+fe+e+++e+r++Gt+vg++d+dp rW++SkWr+Lk
Neem_11185_f_1 276 AWHAVSTGTLFTVYYKPRTSPAEFIVPFDQYRESIKSNYSIGMRFKMRFEGEEAPEQRFTGTIVGIEDADPKRWRESKWRCLK 358
89*******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 3.01E-40 | 146 | 279 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.8E-40 | 147 | 265 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.42E-20 | 148 | 250 | No hit | No description |
| SMART | SM01019 | 4.1E-22 | 150 | 252 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 1.4E-20 | 150 | 251 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.42 | 150 | 252 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.5E-36 | 276 | 358 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 4.6E-10 | 698 | 804 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 25.101 | 716 | 798 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 3.24E-11 | 716 | 793 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 841 aa Download sequence Send to blast |
MAAASSSEVS IKSSNETSRK PMEGKNSNST SSGNKRVLDP EMALYTELWH ACAGPLVTVP 60 REGEHVYYFP QGHIEQVEAS TNQVADQQMP VYDLPSKILC RVINVQLKAE PDTDEVFAQV 120 TLLPETNQDE NAVEKEPPPP PPPRFHVHSF CKTLTASDTS THGGFSVLRR HADECLPPLD 180 MSRQPPTQEL VAKDLHANEW RFRHIFRGQP RRHLLQSGWS VFVSSKRLVA GDAFIFLRGE 240 NGELRVGVRR AMRQQGNVPS SVISSHSMHL GVLATAWHAV STGTLFTVYY KPRTSPAEFI 300 VPFDQYRESI KSNYSIGMRF KMRFEGEEAP EQRFTGTIVG IEDADPKRWR ESKWRCLKVR 360 WDETSTIPRP ERVSPWKIEP ALAPPALNPL PMPRPKRPRS SMVPSSPDSS VLTREGSSKV 420 NVDPSSATVF SRVLQGQEFS TLRGNFAERE SNESDTAEKS VVWPPSLEDE KIDVVSASRR 480 YGSENWGPSG RHEPTYTDLL SGFGANGDSP RGYNSPFADQ SVRKSMLDQE GKFNLLARPW 540 SLMPSGLSLK LSESNSKVPV QGGDVNYQVR GNVRYGGFGD YPVLHGNRSE HSHGNWLMPP 600 LPPAQFENSA HSRELMPKPV LVQDQEAGKS KDCKLFGIPL FSNPVVPEPH RNMVNEPVGN 660 IDQQFRAFES DQKSEQSKSS KLADDNKAVN DREKKLQACQ PHTKDVRSKP QSVSARSCTK 720 VHKQGIALGR SVDLSKFNNY DELIAELDQL FEFGGELMAP KKNWLIVYTD DEDDMMLVGD 780 DPWQEFCGMV RKIYIYTKEE VQKMNPGTLS SRGEENSLIG EGGDVKETKQ SLPTVSNMEN 840 C |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-179 | 43 | 380 | 18 | 354 | Auxin response factor 1 |
| 4ldw_A | 1e-179 | 43 | 380 | 18 | 354 | Auxin response factor 1 |
| 4ldw_B | 1e-179 | 43 | 380 | 18 | 354 | Auxin response factor 1 |
| 4ldx_A | 1e-179 | 43 | 380 | 18 | 354 | Auxin response factor 1 |
| 4ldx_B | 1e-179 | 43 | 380 | 18 | 354 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Could act as transcriptional activator or repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Functions independently of ethylene and cytokinin response pathways. May act as a repressor of cell division and organ growth. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952, ECO:0000269|PubMed:16339187}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
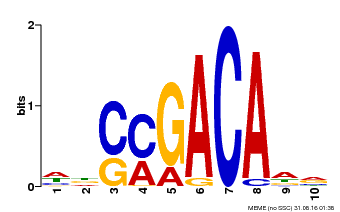
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN600520 | 0.0 | JN600520.1 Citrus sinensis auxin-response factor (ARF) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001275789.1 | 0.0 | auxin-response factor | ||||
| Swissprot | Q94JM3 | 0.0 | ARFB_ARATH; Auxin response factor 2 | ||||
| TrEMBL | G9I820 | 0.0 | G9I820_CITSI; Auxin response factor | ||||
| STRING | XP_006436945.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5348 | 26 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||



