 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_1334_f_14 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1641aa MW: 185150 Da PI: 8.8026 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.4 | 0.00099 | 1525 | 1550 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++sF +k +L+ H r+ +
Neem_1334_f_14 1525 YQCDmeGCTMSFGSKQELVLHKRNiC 1550
99********************9877 PP
| |||||||
| 2 | zf-C2H2 | 12.8 | 0.00037 | 1550 | 1572 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
Neem_1334_f_14 1550 CPvkGCGKKFFSHKYLVQHRRVH 1572
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 11.7 | 0.00081 | 1608 | 1634 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
Neem_1334_f_14 1608 YVCAepGCGQTFRFVSDFSRHKRKtgH 1634
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.5E-14 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.716 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 6.3E-15 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 5.7E-19 | 194 | 334 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 19.123 | 197 | 334 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.59E-13 | 208 | 354 | No hit | No description |
| Pfam | PF02373 | 1.1E-15 | 227 | 278 | IPR003347 | JmjC domain |
| SMART | SM00355 | 9.5 | 1525 | 1547 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 5.31E-6 | 1548 | 1584 | No hit | No description |
| PROSITE profile | PS50157 | 12.362 | 1548 | 1577 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0045 | 1548 | 1572 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.6E-5 | 1549 | 1571 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1550 | 1572 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.6E-9 | 1572 | 1600 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.741 | 1578 | 1607 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0014 | 1578 | 1602 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1580 | 1602 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.24E-9 | 1588 | 1632 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.7E-9 | 1601 | 1631 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.032 | 1608 | 1639 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.62 | 1608 | 1634 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1610 | 1634 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1641 aa Download sequence Send to blast |
MAEAIQQQDI LPWLKTLPSA PEFHPTLAEF QDPIAYIFKI EKEASQYGIC KIVPPVPPAH 60 KKTAITNLNR SLAERAAATG PSSSSSSSSA PTFTTRQQQI GFCPRKPRPV QKPVWQSGEY 120 YTFQQFETKA KNFEKSYLKK CSNKKGPPSA LEIESLYWRA TVDKPFSVEY ANDMPGSAFV 180 PVKKNREVVG EGMTVGETAW NMRGVSRAKG SLLRFMKEEI PGVTSPMVYI AMMFSWFAWH 240 AEDHDLHSLN YLHMGASKTW YGVPMEAANA FEEVVRVHGY GGEINPLGFW LGSLLQILIR 300 LFEFFPLGFN CGEAANIATP EWLSIAKDAA IRRASINYPP MVSHFQLLYD LAIALHSSIP 360 MTISPKPRSS RLKDKKKDEG ETLVKELFVQ DVAQNNELLY ILGKGSPIVL LPESSLDALG 420 ANPRMPHGVC NYREAIKSPG GSVSNDLMIG RGNGVKPLKG FYPMKGKFTS LCERNSSLGG 480 TDNICTWSSQ MLPTDTEREE AGGQADRLSD QRLFSCVTCG ILSFSCVAVI QPREPTARYL 540 MSADCSFFND WIVGSGVTSA GFNVAGGDAV TSEQNSHRRS MGKSGPHSLY DVPVQSIDKI 600 QTVDHSNETI SGRSTERDTS ALNLLAMTYG NSSDSEEDQV EPNVAVCNDE ETNRPKCSLA 660 INYQQDFHGG ASGSLSLPRL DCRDKGPLQT IDCYVEPEFK SENFNDRNPE ISNCSVDFES 720 EKLDSSKANG LNGLLGDTDP ITISHASNCS PAVHGVENVE FSKAIVPIQN ADMSFTPRFD 780 EDSSRMHVFC LEHAVEVEQQ LRPIGGVHIL LLCHPDYPKI VAEAKLVAEE LEIDSFWNDI 840 SFRNATKEDE ERVQLALDSE DAIPGNGDWA VKLGINLFYS ANLSRSPLYS KQMAYNSVIY 900 NAFGRSSPAS SPTKSDYGRR PGRQRKVVAG KWCGKVWMSN QVHPFLSQKD PEEQEQERSF 960 LAWATPDEHL ERKPETTHHI QSTSETRKFS RKRKMPAESM SIKKVKCVDT EDAGSEDSEE 1020 DDTRIQQRRI LRSKPAKFME REDANSYDSP EDNSHVQKRS ASRRKVKCVE REVRVSNDLP 1080 PENSIKQYRR ISKSKQAKCA AREDVVLDDL PDDSSLKQYR KIPRSKLAKY ADRDDEVSDN 1140 SQGDSSYKQL KRIPRSKEAK WIGREDAVSD DSLEDNSRLL QYKRIPKRKQ AKWIEREAAA 1200 SDDSMEDNSQ QLQHKRITKS KKAKCIERED AVSEDSLEDN CRQLQHKRIS KSKQAKRIET 1260 EDVVSDDSLE NKSRQFHRRT PKSKQAEWVE REDAVSDDSL EDNSQQLCRR IPNSKRAKWI 1320 EREDAVSDNS LEDNAHQQHH RITRDKLAKC IEMEDAVSYD SAEDNSHQLR KIPRSKKAKF 1380 TERVDAVSYD SLEDNSLKKR GRILRSKQRK SETLRKMKQE TAWHMKTGKR SLTKQESPQQ 1440 MKQVTRQRSA KGVQNARQFD SFAEEELEGG PSTRLRKRIP KPVKAVVAKS KEKKQTTKKK 1500 VKNASAVKAP AGLSNAKVKD EEAGYQCDME GCTMSFGSKQ ELVLHKRNIC PVKGCGKKFF 1560 SHKYLVQHRR VHLDDRPLKC PWKGCKMTFK WAWARTEHIR VHTGARPYVC AEPGCGQTFR 1620 FVSDFSRHKR KTGHSTKKGR G |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 2e-70 | 1523 | 1638 | 21 | 136 | Lysine-specific demethylase REF6 |
| 6a58_A | 2e-70 | 1523 | 1638 | 21 | 136 | Lysine-specific demethylase REF6 |
| 6a59_A | 2e-70 | 1523 | 1638 | 21 | 136 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 989 | 994 | SRKRKM |
| 2 | 1182 | 1189 | KRIPKRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
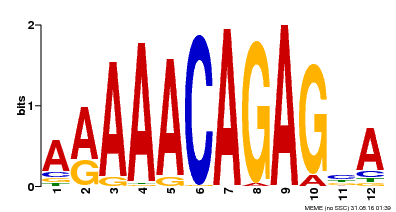
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006476948.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | V4TIR4 | 0.0 | V4TIR4_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006476948.1 | 0.0 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



