 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_14716_f_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 441aa MW: 48696.7 Da PI: 8.5465 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 37.2 | 5.2e-12 | 374 | 419 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L++l P++ +k + a++L +AveYIk+Lq
Neem_14716_f_1 374 SIAERVRRTRISERMRKLQDLFPNL----DKQTNTADMLDLAVEYIKDLQ 419
689*********************9....788*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.532 | 368 | 418 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 4.84E-15 | 371 | 431 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.37E-12 | 372 | 423 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 2.8E-14 | 373 | 431 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.0E-11 | 374 | 424 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 8.7E-9 | 374 | 419 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 441 aa Download sequence Send to blast |
MMYGFAGLQG FPGKMSLLYS HNFKYPEGDL RKNQEFMDSS PHYHQENYNQ QHNSGLTRFR 60 SAPSSFLESL ANGSNSNNSG GGNNCEDFRY FRPSSPEIDT FLAKYMIPSN GSGDSGSHDL 120 LEFGEKTMKQ EESEYSNGSS QMIYQNLPVH SFANDNSVGV GNTVDNSFGV VSSIAMENSM 180 QEKIAAGNGS NNLIRQNSSP AGLFSNLGVD NGYGVMRDVG CFRAGSGTNG ETSTSTSRLN 240 NNINFSSGTS TRFMPQIAEH GNQTIRTSSP EKSLGGNNGS SRQYMSNFTH DTWDSTPFSG 300 IKRVRDNDSN IFFGINASET QNGNSGNQTT RLTHHLSLPK TSAEMAAVEK FLHFQGSVPC 360 KIRAKRGCAT HPRSIAERVR RTRISERMRK LQDLFPNLDK QTNTADMLDL AVEYIKDLQK 420 QVKTLTDNKA KCTCSSKQKQ Y |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00646 | PBM | Transfer from LOC_Os08g39630 | Download |
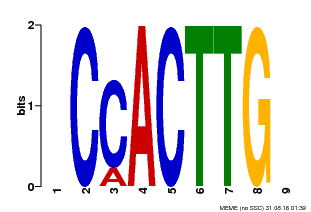
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006482911.1 | 0.0 | transcription factor bHLH130-like | ||||
| Refseq | XP_006482912.1 | 0.0 | transcription factor bHLH130-like | ||||
| TrEMBL | A0A2H5N9Q1 | 0.0 | A0A2H5N9Q1_CITUN; Uncharacterized protein | ||||
| STRING | XP_006482911.1 | 0.0 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM16737 | 8 | 9 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42280.1 | 3e-72 | bHLH family protein | ||||



