 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_26994_f_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 369aa MW: 40673.6 Da PI: 6.9852 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.7 | 4.8e-17 | 45 | 89 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a +++G g W+ I +++g ++t+ q++s+ qk+
Neem_26994_f_1 45 REKWTEEEHQRFLEALRLYGRG-WRQIEEHVG-TKTAVQIRSHAQKF 89
789*******************.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.87E-16 | 39 | 93 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.722 | 40 | 94 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 6.5E-17 | 43 | 92 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.1E-12 | 44 | 92 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.8E-15 | 45 | 88 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-9 | 45 | 85 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.82E-11 | 47 | 90 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 369 aa Download sequence Send to blast |
MSNSSVAAGK NGSQSESAVQ VKELYSFEND PLPKVRKPYT ITKQREKWTE EEHQRFLEAL 60 RLYGRGWRQI EEHVGTKTAV QIRSHAQKFF SKVVRESNGS AESSIMPIEI PPPRPKRKPM 120 HPYPRKHVES LKGTSVSNQQ ERLQSSNALV SDTDRQSPTS ILSAFALDTL GPAVSDQQNG 180 CSSPTSCTTE MHSANLLPIE KEIEYVTSNS SPEEEKVPTL SAQLSAGSNM DHMSVKTESQ 240 SKDSVCIKGD AAVVPTCSSI KLFGTTVLIT DSQKPYLPSA EICKSPKSNC SQENLDIDKK 300 NFVQSVPSKH LDTCLSIGMV ASPSSVGKLV FSHMELQKES ISIAETSPDS YLPFCSNTRR 360 DFEREGNSE |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
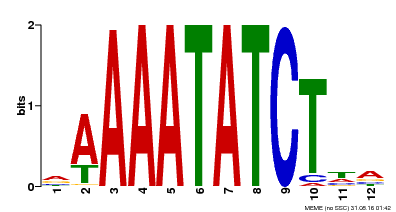
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006478113.1 | 1e-165 | protein REVEILLE 7-like | ||||
| TrEMBL | A0A2H5PXN0 | 1e-164 | A0A2H5PXN0_CITUN; Uncharacterized protein | ||||
| STRING | XP_006478113.1 | 1e-164 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9174 | 27 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.1 | 4e-41 | MYB_related family protein | ||||



