 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_3108_f_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 469aa MW: 51424.7 Da PI: 4.7127 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 37.3 | 5.1e-12 | 304 | 349 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
h+++Er RR++i +++ +L+el+P++ K +Ka++L + ++Y+k Lq
Neem_3108_f_2 304 HSIAERLRREKIAERMKKLQELIPNS-----NKTDKASMLDEIIDYVKFLQ 349
99***********************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 7.07E-17 | 297 | 359 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.89E-13 | 297 | 353 | No hit | No description |
| PROSITE profile | PS50888 | 17.307 | 299 | 348 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-16 | 301 | 358 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.1E-9 | 304 | 349 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.2E-13 | 305 | 354 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 469 aa Download sequence Send to blast |
MDEYIDRCFS SSSWSDANAK ERSSWDYYEP VQQNRVLTNS LEVYEDDRKN SPGSIIRANH 60 IMDSLAVQYT SSLVIGGELE YGIDKNLDFQ EARHERDDQN CNGNLSSSEK MNGSLELGNF 120 GLRYDTAIPT LDSVNLSSPK RLPSVGGMQS SLPFIDGVQV GGNGGELSEV QRSLRDLQTY 180 SPSPELWLPP TYDEVSSLSP VMGQQRIQGF CLQGVNDIDT IGNRYVGMDK ILQFDNLPAS 240 IATKGKQGLQ NHALSCFSAE PQTTTTTVGL PSLQQNASAI STGVSNGTGK PRVRARRGQA 300 TDPHSIAERL RREKIAERMK KLQELIPNSN KTDKASMLDE IIDYVKFLQL QVKVLSMSRL 360 GAAGAVLPLI TDGQAEGANG LSFSPQSDQG VEISLNPDQI AFEEEIVKLM ESNVTMAMQY 420 LQSKGLCLMP IALAAAISSS GKAFEEKHKL GFDWEKTTST FLLKEGLES |
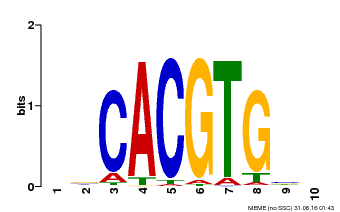
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00659 | PBM | Transfer from Pp3c14_15890 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006474565.1 | 0.0 | uncharacterized protein LOC102611966 isoform X1 | ||||
| Refseq | XP_015384588.1 | 0.0 | uncharacterized protein LOC102611966 isoform X1 | ||||
| TrEMBL | A0A2H5NBL4 | 0.0 | A0A2H5NBL4_CITUN; Uncharacterized protein | ||||
| STRING | XP_006474565.1 | 0.0 | (Citrus sinensis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM16266 | 9 | 14 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58010.1 | 2e-60 | LJRHL1-like 3 | ||||



