 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_3657_f_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 316aa MW: 34060.7 Da PI: 6.2368 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 30.4 | 8.5e-10 | 228 | 276 | 5 | 53 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelk 53
kr++r NR +A rs +RK +i+eLe+kv++L++e ++L +l l+
Neem_3657_f_1 228 KRAKRIWANRQSAARSKERKMRYIAELERKVQTLQTEATSLSAQLTLLQ 276
9*****************************************9998876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 9.8E-16 | 224 | 288 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.932 | 226 | 289 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 7.7E-11 | 228 | 281 | No hit | No description |
| SuperFamily | SSF57959 | 7.74E-11 | 228 | 279 | No hit | No description |
| Pfam | PF00170 | 5.2E-9 | 228 | 276 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14703 | 8.83E-27 | 229 | 278 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 316 aa Download sequence Send to blast |
MDKDKSSSHG GGLPPPSGRF SSFSPPGNHN FNVKPEPSST SSFPPLMPGG SGSDGAHFGH 60 QSDGNRFSHD ISRMPDNPPK NVGHRRAHSE ILTLPDDISF DSDLGVVGGA DGPSFSDETE 120 EDLLSMYLDM DKFNSSTATS GFQMGEPSAA AVPAPPVATG VGNSMVENVG AGSAERPRIR 180 HQHSQSMDGS TSIKPEMLMA GSEESPATDS KKAISAAKLA ELALIDPKRA KRIWANRQSA 240 ARSKERKMRY IAELERKVQT LQTEATSLSA QLTLLQRDTN GLTAENSELK LRLQTMEQQV 300 HLQDDLMDRH TGNHNF |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
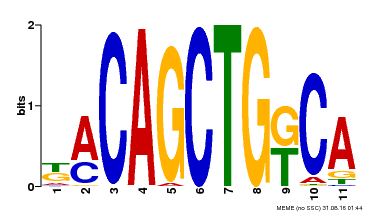
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006442989.1 | 0.0 | probable transcription factor PosF21 | ||||
| Refseq | XP_006478733.1 | 0.0 | probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-131 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | V4VSX8 | 0.0 | V4VSX8_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006478733.1 | 0.0 | (Citrus sinensis) | ||||
| STRING | XP_006442989.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2445 | 27 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-142 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



