 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_4411_f_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 237aa MW: 26317.3 Da PI: 8.2298 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 103.4 | 4.7e-32 | 11 | 123 | 2 | 117 |
YABBY 2 dvfssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaeshldeslkeelleelkveeenlksnvekeesastsvs 97
d+ ssseq+Cyv+CnfC+t+lavsvP tslfk+vtvrCGhCt+llsvn++ a + hl++++ ++ ++ +++ +++ ++++ +++ ++
Neem_4411_f_2 11 DHLSSSEQLCYVHCNFCDTVLAVSVPCTSLFKTVTVRCGHCTNLLSVNMRGLLLPAANQLHLGHAFFTPQNLLEEIRNTPTNMLMINQPNPT--EP 104
7899******************************************9988887777777788888776644333444444445555554444..44 PP
YABBY 98 seklsenedeevprvppvir 117
++ + d+e+p++p v+r
Neem_4411_f_2 105 VMP-VRGIDQEIPKPPVVNR 123
433.4678889998777776 PP
| |||||||
| 2 | YABBY | 121.3 | 1.5e-37 | 146 | 199 | 117 | 170 |
YABBY 117 rPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahfPkihfgl 170
PekrqrvPsaynrfik+eiqrika+nPdishreafsaaaknWahfP+ihfgl
Neem_4411_f_2 146 ETPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGL 199
67**************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 5.4E-65 | 15 | 199 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 1.31E-7 | 144 | 192 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 2.2E-4 | 148 | 191 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 237 aa Download sequence Send to blast |
MSSSTSAFSP DHLSSSEQLC YVHCNFCDTV LAVSVPCTSL FKTVTVRCGH CTNLLSVNMR 60 GLLLPAANQL HLGHAFFTPQ NLLEEIRNTP TNMLMINQPN PTEPVMPVRG IDQEIPKPPV 120 VNRQKLAKAL ACCPPAFTVD IFAFKETPEK RQRVPSAYNR FIKDEIQRIK AGNPDISHRE 180 AFSAAAKNWA HFPHIHFGLM PSDQPVKKTS IRQQEGEDAM NMKDGFFAPA NVGVSPY |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the abaxial cell fate determination during embryogenesis and organogenesis. Regulates the initiation of embryonic shoot apical meristem (SAM) development (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6, PubMed:19837869). Required during flower formation and development, particularly for the patterning of floral organs. Positive regulator of class B (AP3 and PI) activity in whorls 2 and 3. Negative regulator of class B activity in whorl 1 and of SUP activity in whorl 3. Interacts with class A proteins (AP1, AP2 and LUG) to repress class C (AG) activity in whorls 1 and 2. Contributes to the repression of KNOX genes (STM, KNAT1/BP and KNAT2) to avoid ectopic meristems. Binds DNA without sequence specificity. In vitro, can compete and displace the AP1 protein binding to DNA containing CArG box (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6). {ECO:0000269|PubMed:10323860, ECO:0000269|PubMed:10331982, ECO:0000269|PubMed:10457020, ECO:0000269|PubMed:11812777, ECO:0000269|PubMed:12417699, ECO:0000269|PubMed:19837869, ECO:0000269|PubMed:9878633, ECO:0000269|Ref.3, ECO:0000269|Ref.6}. | |||||
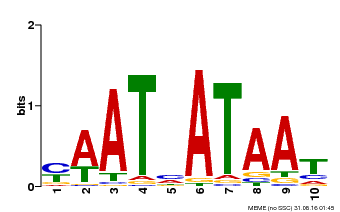
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM468296 | 4e-44 | AM468296.2 Vitis vinifera contig VV78X023826.39, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006445302.1 | 1e-130 | axial regulator YABBY 1 | ||||
| Refseq | XP_006490878.1 | 1e-130 | axial regulator YABBY 1 | ||||
| Swissprot | O22152 | 1e-92 | YAB1_ARATH; Axial regulator YABBY 1 | ||||
| TrEMBL | A0A067H4B3 | 1e-129 | A0A067H4B3_CITSI; Uncharacterized protein | ||||
| TrEMBL | A0A2H5NP36 | 1e-129 | A0A2H5NP36_CITUN; Uncharacterized protein | ||||
| TrEMBL | V4TY63 | 1e-129 | V4TY63_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006490878.1 | 1e-130 | (Citrus sinensis) | ||||
| STRING | XP_006445302.1 | 1e-130 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2217 | 28 | 77 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45190.1 | 6e-95 | YABBY family protein | ||||



