 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_4507_f_4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 675aa MW: 74637.3 Da PI: 8.7148 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52.2 | 1.1e-16 | 345 | 391 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+ksLq
Neem_4507_f_4 345 VHNLSERRRRDRINEKMRALQELIPHC-----NKTDKASMLDEAIEYLKSLQ 391
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.05E-20 | 338 | 403 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.457 | 341 | 390 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.65E-17 | 344 | 395 | No hit | No description |
| Pfam | PF00010 | 5.3E-14 | 345 | 391 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.0E-20 | 345 | 399 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.3E-18 | 347 | 396 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 675 aa Download sequence Send to blast |
MNPCMPDWNF EGDLPVSNQM KSTGTDHELV ELLWRNGQVV LQSQTHRKPT LNPTEPRQVQ 60 KQTIRGNSSS YGNSSNLIQN DETVSWIQYP IEDSLEKEFY SNFFSDLPPS GPMEVDKHAR 120 QLEGEKSVKF DVPGVVTSLQ HSNDNRSAVP GLQRSTMPPP RFEFHDAAQQ NKSLGDLGKV 180 VNFPQSTAAL KGELGSGKGQ IDRKVSGNLP QGEVRECSMM TVGSSHCGSN QVAYDIDMSR 240 ASSNGVGSTG LSPGNLKDDV RKVITQSDRG KTETLEPTVT SSSGGSGSSF NRTSKQSTGD 300 NSLKRKNRDA GESECQSEAA EFESAGGNRT AQRSGSSRRS RAAEVHNLSE RRRRDRINEK 360 MRALQELIPH CNKTDKASML DEAIEYLKSL QLQLQVMWMG SGMAPVMFPG VQHYVSRMGM 420 GMGPPSLPSI TNTMHLPRVP LVDQSMSMAQ AQNQAVMCQT PVLNPVNYQN QMQNSNFSDQ 480 YARYMGFHPM QTTSQPMNMF RFGSPTMQSQ IVSQPSSTCG PFPGGAASDN APLSGKTGRL 540 HLSSQMVMLL FVGQVEDQLL KIAYLAAVSV TFIFIILDGF ILIRTLSRFA VLTLIWSLRS 600 YWPKILAPSL PYSLFHTHRS TMTVNNINCE IHSHALKVQL KATATRHQLL CINHTANSKS 660 CDHPLLRKDF CPQDR |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 349 | 354 | ERRRRD |
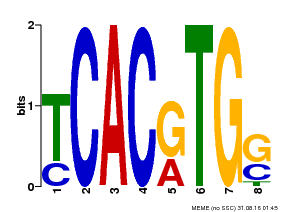
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024042297.1 | 0.0 | transcription factor PIF4 isoform X2 | ||||
| TrEMBL | V4TKJ1 | 0.0 | V4TKJ1_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006437070.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3468 | 25 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G59060.4 | 8e-45 | phytochrome interacting factor 3-like 6 | ||||



