 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_6825_f_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 243aa MW: 27096.3 Da PI: 6.3155 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 58.8 | 1.3e-18 | 95 | 144 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr ++ +g+++AeIrd + ng r++lg+f++ae Aa a+++a+ +++g
Neem_6825_f_1 95 YRGVRKRP-WGKYAAEIRDSTRNG--VRVWLGTFDSAEAAALAYDQAAFSMRG 144
9*******.**********44465..*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 2.3E-37 | 94 | 158 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.2E-13 | 94 | 144 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 5.7E-30 | 94 | 152 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 22.695 | 94 | 152 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.47E-22 | 95 | 154 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 3.3E-10 | 95 | 106 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.98E-20 | 95 | 153 | No hit | No description |
| PRINTS | PR00367 | 3.3E-10 | 118 | 134 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 243 aa Download sequence Send to blast |
MESSFLSYPK PEFSPASSFG SSHDSLSWDE LLLSHNSLPF NINDSEEMLL LGVLAEGTTK 60 LESSESISVD GVNKEEEVTS SSKTVEESSK KERSYRGVRK RPWGKYAAEI RDSTRNGVRV 120 WLGTFDSAEA AALAYDQAAF SMRGSLATLN FPIEMVKESL QEMKYRCEDG CSPVVALKRR 180 HSMRKKSSTV NKKSKKKELI DHHHHHQVQV GSQNVVVLED LGADYLEQLL MSSCESSGAT 240 PNW |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 2e-28 | 93 | 155 | 1 | 63 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the GCC-box cis-acting elements found in the promoter regions of ethylene-responsive genes (PubMed:23057995). Acts downstream of MYC2 in the jasmonate-mediated response to Botrytis cinerea infection (PubMed:28733419). With MYC2 forms a transcription module that regulates pathogen-responsive genes (PubMed:28733419). {ECO:0000269|PubMed:23057995, ECO:0000269|PubMed:28733419}. | |||||
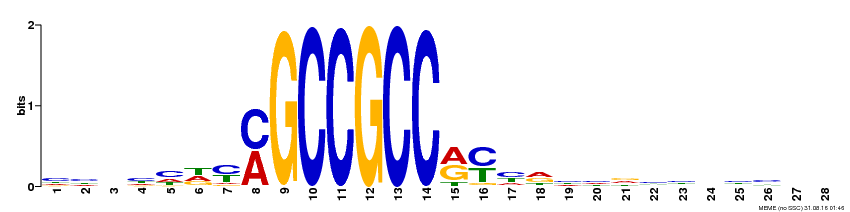
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00374 | DAP | Transfer from AT3G23240 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding and infection with the fungal pathogen Botrytis cinerea. {ECO:0000269|PubMed:28733419}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023906372.1 | 6e-92 | ethylene-responsive transcription factor 1B-like | ||||
| Swissprot | A0A3Q7I5Y9 | 2e-74 | ERFC3_SOLLC; Ethylene-response factor C3 | ||||
| TrEMBL | D8VD37 | 1e-99 | D8VD37_ACTDE; Ethylene response factor 10 | ||||
| STRING | EOX94471 | 2e-85 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2275 | 28 | 76 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23240.1 | 6e-50 | ethylene response factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



