 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_8072_f_3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 596aa MW: 69141.8 Da PI: 9.2501 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 25.6 | 2.7e-08 | 53 | 73 | 5 | 25 |
CHHHCHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKk 25
k err+++NReAAr+sR+RKk
Neem_8072_f_3 53 KTERRLAQNREAARKSRLRKK 73
89******************8 PP
| |||||||
| 2 | bZIP_1 | 22.6 | 2.2e-07 | 92 | 109 | 8 | 25 |
HCHHHHHHHHHHHHHHHH CS
bZIP_1 8 rrkqkNReAArrsRqRKk 25
r+++NReAAr+sR+RKk
Neem_8072_f_3 92 LRLAQNREAARKSRLRKK 109
599**************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 6.2E-5 | 48 | 74 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 56 | 71 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 2.7E-5 | 93 | 145 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 93 | 107 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 4.3E-29 | 189 | 264 | IPR025422 | Transcription factor TGA like domain |
| PROSITE profile | PS50076 | 15.778 | 404 | 478 | IPR001623 | DnaJ domain |
| PRINTS | PR00625 | 2.4E-10 | 409 | 427 | IPR001623 | DnaJ domain |
| SMART | SM00271 | 9.9E-10 | 413 | 470 | IPR001623 | DnaJ domain |
| CDD | cd06257 | 1.05E-11 | 413 | 467 | IPR001623 | DnaJ domain |
| Pfam | PF00226 | 6.3E-16 | 414 | 475 | IPR001623 | DnaJ domain |
| SuperFamily | SSF46565 | 1.01E-17 | 414 | 476 | IPR001623 | DnaJ domain |
| Gene3D | G3DSA:1.10.287.110 | 5.5E-17 | 415 | 477 | IPR001623 | DnaJ domain |
| PRINTS | PR00625 | 2.4E-10 | 427 | 442 | IPR001623 | DnaJ domain |
| PRINTS | PR00625 | 2.4E-10 | 450 | 470 | IPR001623 | DnaJ domain |
| PROSITE pattern | PS00636 | 0 | 455 | 474 | IPR018253 | DnaJ domain, conserved site |
| PRINTS | PR00625 | 2.4E-10 | 496 | 515 | IPR001623 | DnaJ domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 596 aa Download sequence Send to blast |
MSSTSTELAA LRRMSIFEPY PPISMWGDAF QGDASPNTDS STIVQVDARL DNKTERRLAQ 60 NREAARKSRL RKKVLGDAFI LLFLWMEGTH YLRLAQNREA ARKSRLRKKV LGDAFILLFY 120 GWKAYVQQLE SSRLKLAQLE QELERTRHQG LYIGIASDGS HYGLSAHINP GITAFEMEYG 180 HWVEEQNRQI CELRNALQAH ITDIELRILV ESGLNHYYNL FRMKADAAKA DVFYLISGMW 240 RTSTERFFQW IGGFRPSELL NILMPQIEPL TDQQVVDVCN LRQSSQQAED ALSQGIDKLQ 300 QTLVQSLTED QLISGSYHSQ MAAAVEKLDA LESFVNQADH LRQQTMQQMY RILTTRQAAR 360 ALLALGEYFH RLRALIVLPM TCRSYKYQNT VMSFDSLMVE LVGNYRTRNG QNELGRDADE 420 EQIKVAYRRL AKFYHPDVYD GRGTLEEGET AEARFIKIQA AYELLIDEER RRQYDKDNRV 480 NPMKASQAWM QWLIKKRKAF DQRGDMAIAA WAEQQQLELN LRARRLSHSK VDPEEERRIL 540 AREKKASKEY FNNTLKRHTL VLKKRDLMRK KAEEKKRAIS QLLAAEGLEL DTDDEA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
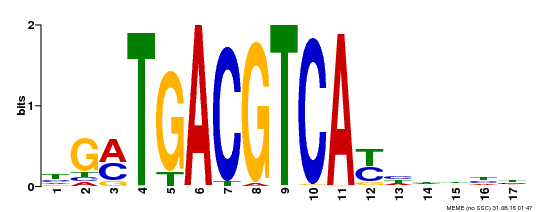
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1 (By similarity). {ECO:0000250, ECO:0000269|PubMed:12953119}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00247 | DAP | Transfer from AT1G77920 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015384214.1 | 0.0 | transcription factor TGA7 isoform X2 | ||||
| Swissprot | Q93ZE2 | 1e-147 | TGA7_ARATH; Transcription factor TGA7 | ||||
| TrEMBL | A0A371FDF4 | 0.0 | A0A371FDF4_MUCPR; Transcription factor TGA3 (Fragment) | ||||
| STRING | XP_006434079.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3061 | 27 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G77920.1 | 1e-139 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



