 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_8133_f_4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 394aa MW: 45703.1 Da PI: 6.7024 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 180.1 | 5.8e-56 | 16 | 144 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgyWkatgkdkevl 93
+ppGfrFhPtdeelv++yL+kk+++k+++l +vik+vd+yk+ePwdL+ k+ + e++ewyfFs++dkky+tg+r+nratk+g+Wkatg+dk+++
Neem_8133_f_4 16 VPPGFRFHPTDEELVDYYLRKKIASKRIDL-DVIKDVDLYKIEPWDLQDlcKIGTeEQNEWYFFSHKDKKYPTGTRTNRATKAGFWKATGRDKAIY 110
69****************************.9***************943455552555************************************* PP
NAM 94 skkgelvglkktLvfykgrapkgektdWvmheyrl 128
s +++l+g++ktLvfykgrap+g+k+dW+mheyr
Neem_8133_f_4 111 S-RHSLIGMRKTLVFYKGRAPNGQKSDWIMHEYRH 144
*.889****************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 9.68E-59 | 11 | 154 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.298 | 16 | 193 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.6E-28 | 17 | 143 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 9.68E-59 | 181 | 193 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 394 aa Download sequence Send to blast |
MQVLTPWDNM NSFSHVPPGF RFHPTDEELV DYYLRKKIAS KRIDLDVIKD VDLYKIEPWD 60 LQDLCKIGTE EQNEWYFFSH KDKKYPTGTR TNRATKAGFW KATGRDKAIY SRHSLIGMRK 120 TLVFYKGRAP NGQKSDWIMH EYRHETNENG TPQAMRTVSL KTFFSPSDFL LKHFYPEIIS 180 KEEGWVVCRV FKKRMATVRK MGEYDSSCWY DEQVSFMPEI DSPGRRMPQP FTPYHLHYPC 240 KQELELQYNM SLHDAFLQLP QLESPKVPQS AASVSCNSVV PFGYDRNNGS TLQSSTLTQE 300 EHTQQAHQQD LNSLYSNNNE QAVDQVTDWR VLDKFVASQL SHEDASKENN YSNAADFNVA 360 EQMNSILANG SKRSEIAQDY ASTSTSSCQI DLWK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 2e-47 | 15 | 143 | 14 | 139 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:16103214, PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:25148240}. | |||||
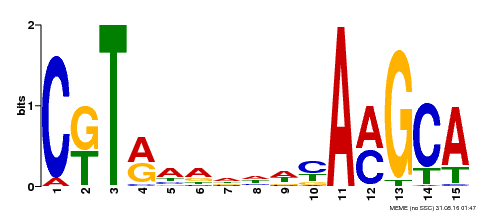
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00136 | DAP | Transfer from AT1G12260 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC775296 | 9e-92 | KC775296.1 Jatropha curcas NAC transcription factor 018 gene, complete cds. | |||
| GenBank | KR605203 | 9e-92 | KR605203.1 Manihot esculenta NAC transcription factors 65 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017973363.1 | 0.0 | PREDICTED: NAC domain-containing protein 7 | ||||
| Swissprot | Q9FWX2 | 1e-158 | NAC7_ARATH; NAC domain-containing protein 7 | ||||
| TrEMBL | A0A061GBJ7 | 0.0 | A0A061GBJ7_THECC; NAC domain protein, IPR003441 isoform 1 | ||||
| STRING | EOY24430 | 0.0 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2111 | 26 | 78 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G12260.1 | 1e-155 | NAC 007 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



